A Practical Framework for Formulating Parameter Estimation Problems in Biomedical Research
This article provides a comprehensive framework for formulating parameter estimation problems, specifically tailored for researchers, scientists, and drug development professionals.
A Practical Framework for Formulating Parameter Estimation Problems in Biomedical Research
Abstract
This article provides a comprehensive framework for formulating parameter estimation problems, specifically tailored for researchers, scientists, and drug development professionals. It guides readers from foundational principles—defining parameters, models, and data requirements—through the core formulation of the problem as an optimization task, detailing objective functions and constraints. The content further addresses advanced troubleshooting, optimization techniques for complex models like pharmacokinetic-pharmacodynamic (PK/PD) relationships, and concludes with robust methods for validating and comparing estimation results to ensure reliability for critical biomedical applications.
Core Concepts: Defining Parameters, Models, and Data for Scientific Inference
What is a Parameter? Distinguishing Between Model Parameters and Population Characteristics
In statistical and scientific research, a parameter is a fundamental concept referring to a numerical value that describes a characteristic of a population or a theoretical model. Unlike variables, which can be measured and can vary from one individual to another, parameters are typically fixed constants, though their true values are often unknown and must be estimated through inference from sample data [1] [2]. Parameters serve as the essential inputs for probability distribution functions and mathematical models, generating specific distribution curves or defining the behavior of dynamic systems [1] [3]. The precise understanding of what a parameter represents—whether a population characteristic or a model coefficient—is critical for formulating accurate parameter estimation problems in research, particularly in fields like drug development and systems biology.
This guide delineates the two primary contexts in which the term "parameter" is used: (1) population parameters, which are descriptive measures of an entire population, and (2) model parameters, which are constants within a mathematical model that define its structure and dynamics. While a population parameter is a characteristic of a real, finite (though potentially large) set of units, a model parameter is part of an abstract mathematical representation of a system, which could be based on first principles or semi-empirical relationships [3] [4]. The process of determining the values of these parameters, known as parameter estimation, constitutes a central inverse problem in scientific research.
Population Parameters
Definition and Key Characteristics
A population parameter is a numerical value that describes a specific characteristic of an entire population [2] [5]. A population, in statistics, refers to the complete set of subjects, items, or entities that share a common characteristic and are the focus of a study [5]. Examples include all inhabitants of a country, all students in a university, or all trees in a forest [5]. Population parameters are typically represented by Greek letters to distinguish them from sample statistics, which are denoted by Latin letters [6] [7].
Key features of population parameters include:
- They Represent Entire Populations: Parameters encapsulate the entire population, whether it comprises millions of individuals or just a handful [5].
- They Are Fixed Constants: A population parameter is a fixed value, unlike a sample statistic, which can vary from one sample to another [1] [5]. This value does not change unless the population itself changes [5].
- They Are Often Unknown: For most real-world populations, especially large ones, measuring every unit is infeasible, making the true parameter value unknown [1] [2]. Researchers must then rely on statistical inference from samples to estimate population parameters.
Common Types of Population Parameters
Population parameters are classified based on the type of data they describe and the aspect of the population they summarize [5].
Table 1: Common Types of Population Parameters
| Type | Description | Common Examples |
|---|---|---|
| Location Parameters | Describe the central point or typical value in a population distribution. | Mean (μ), Median, Mode [2] [5] |
| Dispersion Parameters | Quantify the spread or variability of values around the center. | Variance (σ²), Standard Deviation (σ), Range [5] |
| Proportion Parameters | Represent the fraction of the population possessing a certain characteristic. | Proportion (P) [6] [5] |
| Shape Parameters | Describe the form of the population distribution. | Skewness, Kurtosis [5] |
Population Parameters vs. Sample Statistics
The distinction between a parameter and a statistic is fundamental to statistical inference. A sample statistic (or parameter estimate) is a numerical value describing a characteristic of a sample—a subset of the population—and is used to make inferences about the unknown population parameter [1] [6] [7]. For example, the average income for a sample drawn from the U.S. is a sample statistic, while the average income for the entire United States is a population parameter [6].
Table 2: Parameter vs. Statistic Comparison
| Aspect | Parameter | Statistic |
|---|---|---|
| Definition | Describes a population [6] [5] | Describes a sample [6] [5] |
| Scope | Entire population [5] | Subset of the population [5] |
| Calculation | Generally impractical, often unknown [5] | Directly calculated from sample data [5] |
| Variability | Fixed value [1] [5] | Variable, depends on the sample drawn [1] [5] |
| Notation (e.g., Mean) | μ (Greek letters) [6] [7] | x̄ (Latin letters) [6] [7] |
The relationship between a statistic and a parameter is encapsulated in the concept of a sampling distribution, which is the probability distribution of a given statistic (like the sample mean) obtained from a large number of samples drawn from the same population [1]. This distribution enables researchers to draw conclusions about the corresponding population parameter and quantify the uncertainty in their estimates [1].
Model Parameters
Definition and Role in Mathematical Modeling
In the context of mathematical modeling, a model parameter is a constant that defines the structure and dynamics of a system described by a set of equations [3]. These parameters are not merely descriptive statistics of a population but are integral components of a theoretical framework designed to represent the behavior of a physical, biological, or economic system. Model parameters often represent physiological quantities, physical constants, or system gains and time scales [3]. For instance, in a model predicting heart rate regulation, parameters might represent afferent baroreflex gain or sympathetic delay [3].
The process of parameter estimation involves solving the inverse problem: given a model structure and a set of observational data, predict the values of the model parameters that best explain the data [3]. This is a central challenge in many scientific disciplines, as models can become complex with many parameters, while available data are often sparse or noisy [3].
The Parameter Estimation Problem
Formally, a dynamic system model can often be described by a system of differential equations:
dx/dt = f(t, x; θ)
where t is time, x is the state vector, and θ is the parameter vector [3]. The model output y that corresponds to measurable data is given by:
y = g(t, x; θ) [3].
Given a set of observed data Y sampled at specific times, the goal is to find the parameter vector θ that minimizes the difference between the model output y and the observed data Y [3]. This is typically framed as an optimization problem, where an objective function (often a least squares error) is minimized.
A significant challenge in this process is parameter identifiability—determining whether it is possible to uniquely estimate a parameter's value given the model and the available data [3]. A parameter may be non-identifiable due to the model's structure or because the available data are insufficient to inform the parameter. This leads to the need for subset selection, the process of identifying which parameter subsets can be reliably estimated given the model and a specific dataset [3].
Formulating a Parameter Estimation Problem: A Methodological Framework
Foundational Steps
Formulating a robust parameter estimation problem is a critical step in data-driven research. The following workflow outlines the core process, integrating both population and model parameter contexts.
Diagram: Parameter Estimation Workflow. This flowchart outlines the key stages in formulating and solving a parameter estimation problem for research.
Define the Population and Characteristic of Interest
The first step is to unambiguously define the target population—the entire set of units (people, objects, transactions) about which inference is desired [5] [7]. This involves specifying the content, units, extent, and time. For example, a population might be "all patients diagnosed with stage 2 hypertension in the United States during the calendar year 2024." The population parameter of interest (e.g., mean systolic blood pressure, proportion responding to a drug) must also be clearly defined [8].
Select or Formulate a Mathematical Model
Based on the underlying scientific principles, a mathematical model must be selected or developed. This model, often a system of differential or algebraic equations, represents the hypothesized mechanisms governing the system [3] [4]. The model should be complex enough to capture essential dynamics but simple enough to allow for parameter identification given the available data.
Identify Model Parameters for Estimation
Not all parameters in a complex model can be estimated from a given dataset. Methods for practical parameter identification are used to determine a subset of parameters that can be estimated reliably [3]. Three methods compared in recent research include:
- Structured analysis of the correlation matrix: Identifies highly correlated parameters to avoid estimating them simultaneously. This method can provide the "best" subset but is computationally intensive [3].
- Singular value decomposition followed by QR factorization: A numerical approach to select a subset of identifiable parameters. This method is computationally easier but may sometimes result in subsets that still contain correlated parameters [3].
- Subspace identification: Identifies the parameter subspace closest to the one spanned by eigenvectors of the model Hessian matrix [3].
Experimental and Sampling Design
Design Sampling Strategy or Experiment
For population parameters, this involves designing a sampling plan to collect data from a representative subset of the population. The key is to minimize sampling error (error due to observing a sample rather than the whole population) and non-sampling errors (e.g., measurement error) [5]. For model parameters, this involves designing experiments that will generate data informative for the parameters of interest, often requiring perturbation of the system to excite the relevant dynamics [3].
Collect Data
Data is collected according to the designed strategy. For sample surveys, this involves measuring the selected sample units. For experimental models, this involves measuring the system outputs (the y vector) in response to controlled inputs [3] [8]. The data collection process must be rigorously documented and controlled to ensure quality.
Estimation and Validation
Choose an Estimation Method
The appropriate estimation method depends on the model's nature (linear/nonlinear), the error structure, and the data available.
- For nonlinear models: Local methods (e.g., gradient-based) can be efficient but may converge to local minima. Global optimization methods (e.g., branch-and-bound frameworks) are designed to find the global solution of nonconvex problems but are computationally more demanding [4].
- Error-in-variables methods: Account for noise in both input and output measurements, leading to a more complex but often more realistic formulation [4].
- Bayesian methods: Incorporate prior knowledge about parameters and provide a posterior distribution rather than a single point estimate.
Solve the Inverse Problem
This is the computational core of the process, where the optimization algorithm is applied to find the parameter values that minimize the difference between the model output and the observed data [3] [4]. For dynamic systems with process and measurement noise, specialized algorithms that reparameterize the unknown noise covariances have been developed to overcome theoretical and practical difficulties [9].
Validate and Interpret Results
The estimated parameters must be validated for reliability and interpreted in the context of the research question. This involves:
- Uncertainty Quantification: Calculating confidence intervals or credible regions for the parameter estimates [1] [5].
- Model Validation: Testing the model's predictive performance with the estimated parameters on a new, unseen dataset.
- Sensitivity Analysis: Determining how changes in parameters affect the model output, which also informs the reliability of the estimates [3].
Essential Reagents and Computational Tools for Parameter Estimation
Successful parameter estimation, particularly in biological and drug development contexts, relies on a suite of methodological reagents and computational tools.
Table 3: Research Reagent Solutions for Parameter Estimation
| Category / Tool | Function in Parameter Estimation |
|---|---|
| Sensitivity & Identifiability Analysis | Determines which parameters significantly influence model outputs and can be uniquely estimated from available data [3]. |
| Global Optimization Algorithms (e.g., αBB) | Solves nonconvex optimization problems to find the global minimum of the objective function, avoiding local solutions [4]. |
| Structured Correlation Analysis | Identifies and eliminates correlated parameters to ensure a numerically well-posed estimation problem [3]. |
| Error-in-Variables Formulations | Accounts for measurement errors in both independent and dependent variables, leading to less biased parameter estimates [4]. |
| Sampling Design Frameworks | Plans data collection strategies to maximize the information content for the parameters of interest while minimizing cost [5]. |
| Markov Chain Monte Carlo (MCMC) | For Bayesian estimation, samples from the posterior distribution of parameters, providing full uncertainty characterization. |
Understanding the dual nature of parameters—as fixed population characteristics and as tuning elements within mathematical models—is foundational to scientific research. Formulating a parameter estimation problem requires a systematic approach that begins with a precise definition of the population or system of interest, proceeds through careful model selection and experimental design, and culminates in the application of robust computational methods to solve the inverse problem. The challenges of practical identifiability, correlation between parameters, and the potential for multiple local optima necessitate a sophisticated toolkit. By rigorously applying the methodologies outlined—from subset selection techniques like structured correlation analysis to global optimization frameworks—researchers and drug development professionals can reliably estimate parameters, thereby transforming models into powerful, patient-specific tools for prediction and insight.
Mathematical models serve as a critical backbone in scientific research and drug development, providing a quantitative framework to describe, predict, and optimize system behaviors. The progression from simple statistical models to complex mechanistic representations marks a significant evolution in how researchers approach problem-solving across diverse fields. In pharmaceutical development particularly, this modeling continuum enables more informed decision-making from early discovery through clinical stages and into post-market monitoring [10].
The formulation of accurate parameter estimation problems stands as a cornerstone of effective modeling, bridging the gap between theoretical constructs and experimental data. This technical guide explores the spectrum of mathematical modeling approaches, with particular emphasis on parameter estimation methodologies that ensure models remain both scientifically rigorous and practically useful within drug development pipelines. As models grow in complexity to encompass physiological detail and biological mechanisms, the challenges of parameter identifiability and estimation become increasingly pronounced, especially when working with limited experimental data [3] [11].
The Modeling Spectrum: From Descriptive to Mechanistic
Simple Regression and Statistical Models
The foundation of mathematical modeling begins with statistical approaches that describe relationships between variables without necessarily invoking biological mechanisms. Simple linear regression models, utilizing techniques such as ordinary least squares, establish quantitative relationships between independent and dependent variables [12]. These models provide valuable initial insights, particularly when the underlying system mechanisms are poorly understood.
In pharmacological contexts, Non-compartmental Analysis (NCA) represents a model-independent approach that estimates key drug exposure parameters directly from concentration-time data [10]. Similarly, Exposure-Response (ER) analysis quantifies relationships between drug exposure levels and their corresponding effectiveness or adverse effects, serving as a crucial bridge between pharmacokinetics and pharmacodynamics [10].
Intermediate Complexity: Semi-Mechanistic Models
Semi-mechanistic models incorporate elements of biological understanding while maintaining empirical components where knowledge remains incomplete. Population Pharmacokinetic (PPK) models characterize drug concentration time courses in diverse patient populations, explaining variability through covariates such as age, weight, or renal function [10]. These models employ mixed-effects statistical approaches to distinguish between population-level trends and individual variations.
Semi-Mechanistic PK/PD models combine mechanistic elements describing drug disposition with empirical components capturing pharmacological effects [10]. This hybrid approach balances biological plausibility with practical estimability, often serving as a workhorse in clinical pharmacology applications.
Complex Mechanistic Models
At the most sophisticated end of the spectrum, mechanistic models attempt to capture the underlying biological processes governing system behavior. Physiologically Based Pharmacokinetic (PBPK) models incorporate anatomical, physiological, and biochemical parameters to predict drug absorption, distribution, metabolism, and excretion across different tissues and organs [10] [13]. These models facilitate species translation and prediction of drug-drug interactions.
Quantitative Systems Pharmacology (QSP) models represent the most comprehensive approach, integrating systems biology with pharmacology to simulate drug effects across multiple biological scales [10]. QSP models capture complex network interactions, pathway dynamics, and feedback mechanisms, making them particularly valuable for exploring therapeutic interventions in complex diseases.
Table 1: Classification of Mathematical Models in Drug Development
| Model Category | Key Examples | Primary Applications | Data Requirements |
|---|---|---|---|
| Simple Statistical | Linear Regression, Non-compartmental Analysis (NCA), Exposure-Response | Initial data exploration, descriptive analysis, preliminary trend identification | Limited, often aggregate data |
| Semi-Mechanistic | Population PK, Semi-mechanistic PK/PD, Model-Based Meta-Analysis (MBMA) | Clinical trial optimization, dose selection, covariate effect quantification | Rich individual-level data, sparse sampling designs |
| Complex Mechanistic | PBPK, QSP, Intact Protein PK/PD (iPK/PD) | First-in-human dose prediction, species translation, target validation, biomarker strategy | Extensive in vitro and in vivo data, system-specific parameters |
Parameter Estimation Fundamentals
The Parameter Estimation Problem
Parameter estimation constitutes the process of determining values for model parameters that best explain observed experimental data. Formally, this involves identifying parameter vector θ that minimizes the difference between model predictions y(t,θ) and experimental observations Y(t) [3]. The problem can be represented as finding θ that minimizes the objective function:
[ \min{\theta} \sum{i=1}^{N} [Y(ti) - y(ti,\theta)]^2 ]
where N represents the number of observations, Y(ti) denotes measured data at time ti, and y(ti,θ) represents model predictions at time ti given parameters θ [3].
Practical Identifiability Challenges
A fundamental challenge in parameter estimation lies in determining whether parameters can be uniquely identified from available data. Structural identifiability addresses whether parameters could theoretically be identified given perfect, noise-free data, while practical identifiability considers whether parameters can be reliably estimated from real, noisy, and limited datasets [3]. As model complexity increases, the risk of non-identifiability grows substantially, particularly when working with sparse data common in clinical and preclinical studies.
The heart of the estimation challenge emerges from the inverse problem nature of parameter estimation: deducing internal system parameters from external observations [3]. This problem frequently lacks unique solutions, especially when models contain numerous parameters or when data fails to adequately capture system dynamics across relevant timescales and conditions.
Methodologies for Parameter Estimation
Subset Selection Methods
Subset selection approaches systematically identify which parameters can be reliably estimated from available data while fixing remaining parameters at prior values. These methods rank parameters from most to least estimable, preventing overfitting by reducing the effective parameter space [3] [11]. Three prominent techniques include:
Structured Correlation Analysis: Examines parameter correlation matrices to identify highly correlated parameter groups that cannot be independently estimated [3]. This method provides comprehensive insights but can be computationally intensive.
Singular Value Decomposition (SVD) with QR Factorization: Uses matrix decomposition techniques to identify parameter subsets that maximize independence and estimability [3]. This approach offers computational advantages while providing reasonable subset selections.
Hessian-based Subspace Identification: Analyzes the Hessian matrix (matrix of second-order partial derivatives) to identify parameter directions most informed by available data [3]. This method connects parameter estimability to model sensitivity.
Subset selection proves particularly valuable when working with complex mechanistic models containing more parameters than can be supported by available data [11]. The methodology provides explicit guidance on which parameters should be prioritized during estimation, effectively balancing model complexity with information content.
Bayesian Estimation Methods
Bayesian approaches treat parameters as random variables with probability distributions representing uncertainty. These methods combine prior knowledge (encoded in prior distributions) with experimental data (incorporated through likelihood functions) to generate posterior parameter distributions [11]. This framework naturally handles parameter uncertainty, especially valuable when data is limited.
Bayesian methods introduce regularization through prior distributions, preventing parameter estimates from straying into biologically implausible ranges unless strongly supported by data [11]. This approach proves particularly powerful when incorporating historical data or domain expertise, though it requires careful specification of prior distributions to avoid introducing bias.
Comparison of Estimation Approaches
Table 2: Comparison of Parameter Estimation Methodologies
| Characteristic | Subset Selection | Bayesian Estimation |
|---|---|---|
| Philosophy | Identify and estimate only identifiable parameters | Estimate all parameters with uncertainty quantification |
| Prior Knowledge | Used to initialize fixed parameters | Formally encoded in prior distributions |
| Computational Demand | Moderate to high (multiple analyses required) | High (often requires MCMC sampling) |
| Output | Point estimates for subset parameters | Posterior distributions for all parameters |
| Strengths | Prevents overfitting, provides estimability assessment | Naturally handles uncertainty, incorporates prior knowledge |
| Weaknesses | May discard information about non-estimable parameters | Sensitive to prior misspecification, computationally intensive |
Experimental Protocols for Parameter Estimation
Protocol for Subset Selection Implementation
Model Definition: Formulate the mathematical model structure, specifying state variables, parameters, and input-output relationships [3].
Sensitivity Analysis: Calculate sensitivity coefficients describing how model outputs change with parameter variations. Numerical approximations can be used for complex models: [ S{ij} = \frac{\partial yi}{\partial \thetaj} \approx \frac{y(ti,\thetaj+\Delta\thetaj) - y(ti,\thetaj)}{\Delta\theta_j} ]
Parameter Ranking: Apply subset selection methods (correlation analysis, SVD, or Hessian-based approaches) to rank parameters from most to least estimable [3].
Subset Determination: Select the parameter subset for estimation by identifying the point where additional parameters provide diminishing returns in model fit improvement.
Estimation and Validation: Estimate selected parameters using optimization algorithms, then validate model performance with test datasets [3].
Protocol for Bayesian Estimation Implementation
Prior Specification: Define prior distributions for all model parameters based on literature values, expert opinion, or preliminary experiments [11].
Likelihood Definition: Formulate the likelihood function describing the probability of observing experimental data given parameter values, typically assuming normally distributed errors.
Posterior Sampling: Implement Markov Chain Monte Carlo (MCMC) sampling to generate samples from the posterior parameter distribution [11].
Convergence Assessment: Monitor MCMC chains for convergence using diagnostic statistics (Gelman-Rubin statistic, trace plots, autocorrelation).
Posterior Analysis: Summarize posterior distributions through means, medians, credible intervals, and marginal distributions to inform decision-making.
Research Reagent Solutions for PK/PD Modeling
Table 3: Essential Research Reagents and Materials for PK/PD Studies
| Reagent/Material | Function/Application | Technical Considerations |
|---|---|---|
| Liquid Chromatography Mass Spectrometry (LC-MS/MS) | Quantification of drug concentrations in biological matrices | Provides sensitivity and specificity for drug measurement; essential for traditional PK studies [14] |
| Intact Protein Mass Spectrometry | Measurement of drug-target covalent conjugation (% target engagement) | Critical for covalent drugs where concentration-effect relationship is uncoupled; enables direct target engagement quantification [14] |
| Covalent Drug Libraries | Screening and optimization of irreversible inhibitors | Includes diverse electrophiles for target identification; requires careful selectivity assessment [14] |
| Stable Isotope-Labeled Standards | Internal standards for mass spectrometry quantification | Improves assay precision and accuracy through isotope dilution methods [14] |
| Target Protein Preparations | In vitro assessment of drug-target interactions | Purified proteins for mechanism confirmation and binding affinity determination [14] |
| Biological Matrices | Preclinical and clinical sample analysis | Plasma, blood, tissue homogenates for protein binding and distribution studies [14] |
Visualization of Modeling Workflows
Model-Informed Drug Development Strategy
Parameter Estimation Decision Framework
Covalent Drug Development Workflow
The strategic application of mathematical models across the complexity spectrum provides powerful capabilities for advancing drug development and therapeutic optimization. From simple regression to complex mechanistic PK/PD models, each approach offers distinct advantages tailored to specific research questions and data availability. The critical bridge between model formulation and practical application lies in robust parameter estimation methodologies that respect both mathematical principles and biological realities.
Subset selection and Bayesian estimation approaches offer complementary pathways to address the fundamental challenge of parameter identifiability in limited data environments. Subset selection provides a conservative framework that explicitly acknowledges information limitations, while Bayesian methods fully leverage prior knowledge with formal uncertainty quantification. The choice between these methodologies depends on multiple factors, including data quality, prior knowledge reliability, computational resources, and model purpose.
As Model-Informed Drug Development continues to gain prominence in regulatory decision-making [10], the thoughtful integration of appropriate mathematical models with careful parameter estimation will remain essential for maximizing development efficiency and accelerating patient access to novel therapies. Future advances will likely incorporate emerging technologies such as artificial intelligence and machine learning to enhance both model development and parameter estimation, particularly for complex biological systems where traditional methods face limitations.
The reliability of any parameter estimation problem in scientific research is fundamentally dependent on the triumvirate of data quality, data quantity, and data types. These three elements form the foundational pillars that determine whether mathematical models and statistical analyses can yield accurate, precise, and biologically or chemically meaningful parameter estimates. In model-informed drug development (MIDD) and chemical process engineering, the systematic approach to data collection has become increasingly critical for reducing costly late-stage failures and accelerating hypothesis testing [10] [15]. The framework for reliable estimation begins with recognizing that data must be fit-for-purpose—a concept emphasizing that data collection strategies must be closely aligned with the specific questions of interest and the context in which the resulting model will be used [10].
Parameter estimation represents the process of determining values for unknown parameters in mathematical models that best explain observed experimental data. The precision and accuracy of these estimates directly impact the predictive capability of models across diverse applications, from pharmacokinetic-pharmacodynamic modeling in drug development to process optimization in chemical engineering [10] [15]. This technical guide examines the core requirements for experimental data to support reliable parameter estimation, addressing the interconnected dimensions of quality, quantity, and type that researchers must balance throughout experimental design and execution.
Data Quality Dimensions and Metrics
High-quality data serves as the bedrock of reliable parameter estimation. Data quality can be systematically evaluated through multiple dimensions, each representing a specific attribute that contributes to the overall fitness for use of the data in parameter estimation problems. The table below summarizes the six core dimensions of data quality most relevant to experimental research.
Table 1: Core Data Quality Dimensions for Experimental Research
| Dimension | Definition | Impact on Parameter Estimation | Example Metric |
|---|---|---|---|
| Accuracy | Degree to which data correctly represents the real-world values or events it depicts [16] [17] | Inaccurate data leads to biased parameter estimates and compromised model predictability | Percentage of values matching verified sources [16] |
| Completeness | Extent to which all required data is present and available [16] [18] | Missing data points can introduce uncertainty and reduce estimation precision | Number of empty values in critical fields [18] |
| Consistency | Uniformity of data across different datasets, systems, and time periods [16] [17] | Inconsistent data creates internal contradictions that undermine model identifiability | Percent of matched values across duplicate records [17] |
| Timeliness | Availability of data when needed and with appropriate recency [16] [17] | Outdated data may not represent current system behavior, leading to irrelevant parameters | Time between data collection and availability for analysis [17] |
| Uniqueness | Absence of duplicate or redundant records in datasets [16] [18] | Duplicate records can improperly weight certain observations, skewing parameter estimates | Percentage of duplicate records in a dataset [18] |
| Validity | Conformance of data to required formats, standards, and business rules [16] [17] | Invalid data formats disrupt analytical pipelines and can lead to processing errors | Number of values conforming to predefined syntax rules [17] |
Beyond these core dimensions, additional quality considerations include integrity—which ensures that relationships between data attributes are maintained as data transforms across systems—and freshness, which is particularly critical for time-sensitive processes [17]. For parameter estimation, the relationship between data quality dimensions and their practical measurement can be visualized as a systematic workflow.
The implementation of systematic data quality assessment requires both technical and procedural approaches. Technically, researchers should establish automated validation checks that monitor quality metrics continuously throughout data collection processes [18]. Procedurally, teams should maintain clear documentation of quality standards and assign accountability for data quality management [18]. The cost of poor data quality manifests in multiple dimensions, including wasted resources, unreliable analytics, and compromised decision-making—with one Gartner estimate suggesting poor data quality can result in additional spend of $15M in average annual costs for organizations [17].
Data Types and Their Analytical Implications
Understanding data types is fundamental to selecting appropriate estimation techniques and analytical approaches. Data can be fundamentally categorized as quantitative or categorical, with each category containing specific subtypes that determine permissible mathematical operations and statistical treatments.
Table 2: Data Types in Experimental Research
| Category | Type | Definition | Examples | Permissible Operations | Statistical Methods |
|---|---|---|---|---|---|
| Quantitative | Continuous | Measurable quantities representing continuous values [19] [20] | Height, weight, temperature, concentration [19] | Addition, subtraction, multiplication, division | Regression, correlation, t-tests, ANOVA |
| Discrete | Countable numerical values representing distinct items [19] [20] | Number of patients, cell counts, molecules [19] | Counting, summation, subtraction | Poisson regression, chi-square tests | |
| Categorical | Nominal | Groupings without inherent order or ranking [19] [20] | Species, gender, brand, material type [19] | Equality testing, grouping | Mode, chi-square tests, logistic regression |
| Ordinal | Groupings with meaningful sequence or hierarchy [19] [20] | Severity stages, satisfaction ratings, performance levels [19] | Comparison, ranking | Median, percentile, non-parametric tests | |
| Binary | Special case of nominal with only two categories [20] | Success/failure, present/absent, yes/no [20] | Equality testing, grouping | Proportion tests, binomial tests |
The selection of variables for measurement should be guided by their role in the experimental framework. Independent variables (treatment variables) are those manipulated by researchers to affect outcomes, while dependent variables (response variables) represent the outcome measurements [20]. Control variables are held constant throughout experimentation to isolate the effect of independent variables, while confounding variables represent extraneous factors that may obscure the relationship between independent and dependent variables if not properly accounted for [20].
The relationship between these variable types in an experimental context can be visualized as follows:
In parameter estimation, the data type directly influences the choice of estimation algorithm and model structure. Continuous data typically supports regression-based approaches and ordinary least squares estimation, while categorical data often requires generalized linear models or maximum likelihood estimation with appropriate link functions [20]. The transformation of data types during analysis, such as converting continuous measurements to categorical groupings or deriving composite variables from multiple measurements, should be performed with careful consideration of the information loss and analytical implications [20].
Data Quantity and Experimental Design
Determining the appropriate quantity of data represents a critical balance between statistical rigor and practical constraints. Insufficient data leads to underpowered studies incapable of detecting biologically relevant effects, while excessive data collection wastes resources and may expose subjects to unnecessary risk [21] [22].
Fundamental Principles of Experimental Design
Robust experimental design rests on several key principles that maximize information content while controlling for variability:
- Replication: Repeated observations under identical conditions increase reliability and quantify variability [21]. The number of replicates directly impacts the precision of parameter estimates and should be determined through power analysis rather than historical precedent [22].
- Randomization: Random assignment of treatments to experimental units ensures that unspecified disturbances are spread evenly among treatment groups, preventing confounding [21].
- Blocking: Grouping experimental units into homogeneous blocks allows researchers to remove known sources of variability, thereby increasing the precision of parameter estimates [21].
- Multifactorial Design: Simultaneously varying multiple factors rather than using one-factor-at-a-time approaches enables efficient exploration of factor interactions and more comprehensive parameter estimation [21].
Sample Size Determination through Power Analysis
Statistical power analysis provides a formal framework for determining sample sizes needed to detect effects of interest while controlling error rates. The power (1-β) of a statistical test represents the probability of correctly rejecting a false null hypothesis—that is, detecting an effect when one truly exists [21] [22]. The following diagram illustrates the relationship between key concepts in hypothesis testing and their interconnections:
The parameters involved in sample size calculation for t-tests include:
- Effect Size: The minimum difference between groups that would be considered biologically relevant [22]. For standardized effect sizes, Cohen's d of 0.5, 1.0, and 1.5 typically represent small, medium, and large effects in laboratory animal studies [22].
- Variability: The standard deviation of measurements, typically estimated from pilot studies, previous literature, or systematic reviews [22].
- Significance Level (α): The probability of obtaining a false positive, conventionally set at 0.05 [22].
- Power (1-β): The target probability of detecting a true effect, typically set between 80-95% [22].
For model-based design of experiments (MBDoE) in process engineering and drug development, additional considerations include parameter identifiability and model structure [15]. MBDoE techniques explicitly account for the mathematical model form when designing experiments to maximize parameter precision while minimizing experimental effort [15].
Group Allocation and Balanced Designs
In comparative experiments, proper allocation of experimental units to treatment and control groups is essential. Every experiment should include at least one control or comparator group, which may be negative controls (placebo or sham treatment) or positive controls to verify detection capability [22]. Balanced designs with equal group sizes typically maximize sensitivity for most experimental configurations, though unequal allocation may be advantageous when multiple treatment groups are compared to a common control [22].
Experimental Protocols for Data Generation
Model-Informed Drug Development (MIDD) Protocol
MIDD employs quantitative modeling and simulation to enhance decision-making throughout drug development [10]. The protocol involves:
- Question Formulation: Define specific questions of interest and context of use for the modeling exercise [10].
- Model Selection: Choose appropriate modeling methodologies based on development stage and research questions [10].
- Experimental Design: Identify data requirements and design experiments to inform model parameters [10].
- Data Collection: Execute experiments with appropriate quality controls [10].
- Model Calibration: Estimate parameters using collected data [10].
- Model Validation: Assess model performance against independent data [10].
- Decision Support: Use model for simulation and prediction to inform development decisions [10].
Model-Based Design of Experiments (MBDoE) Protocol
MBDoE represents a systematic approach for designing experiments specifically for mathematical model calibration [15]:
- Model Structure Definition: Establish mathematical model form with unknown parameters [15].
- Parameter Sensitivity Analysis: Identify parameters with greatest influence on model outputs [15].
- Optimal Experimental Design: Determine experimental conditions that maximize information content for parameter estimation [15].
- Experiment Execution: Conduct experiments according to designed conditions [15].
- Parameter Estimation: Fit model parameters to experimental data [15].
- Model Adequacy Assessment: Evaluate model fit and determine if additional experiments are needed [15].
Table 3: Research Reagent Solutions for Experimental Data Generation
| Reagent/Material | Function | Application Examples |
|---|---|---|
| Power Analysis Software | Calculates required sample sizes based on effect size, variability, and power parameters [22] | Determining group sizes for animal studies, clinical trials, in vitro experiments |
| Data Quality Assessment Tools | Automates data validation, completeness checks, and quality monitoring [16] [18] | Continuous data quality assessment during high-throughput screening, clinical data collection |
| Statistical Computing Environments | Implements statistical tests, parameter estimation algorithms, and model fitting procedures [22] | R, Python, SAS for parameter estimation, model calibration, and simulation |
| Laboratory Information Management Systems (LIMS) | Tracks samples, experimental conditions, and results with audit trails [17] | Maintaining data integrity and provenance in chemical and biological experiments |
| Physiologically Based Pharmacokinetic (PBPK) Modeling Software | Simulates drug absorption, distribution, metabolism, and excretion [10] | Predicting human pharmacokinetics from preclinical data, drug-drug interaction studies |
| Quantitative Systems Pharmacology (QSP) Platforms | Integrates systems biology with pharmacology to model drug effects [10] | Mechanism-based prediction of drug efficacy and safety in complex biological systems |
The reliable estimation of parameters in scientific research depends on a systematic approach to data quality, data types, and data quantity. By implementing rigorous data quality dimensions, researchers can ensure that their datasets accurately represent the underlying biological or chemical phenomena under investigation. Through appropriate categorization and handling of different data types, analysts can select estimation methods that respect the mathematical properties of their measurements. Finally, by applying principled experimental design and power analysis, scientists can determine the data quantities necessary to achieve sufficient precision while utilizing resources efficiently.
The integration of these three elements—quality, type, and quantity—creates a foundation for parameter estimation that yields reproducible, biologically relevant, and statistically sound results. As model-informed approaches continue to gain prominence across scientific disciplines, the thoughtful consideration of data requirements at the experimental design stage becomes increasingly critical for generating knowledge that advances both fundamental understanding and applied technologies.
In statistical inference, the estimation of unknown population parameters from sample data is foundational. Two primary paradigms exist: point estimation, which provides a single "best guess" value [23] [24], and interval estimation, which provides a range of plausible values along with a stated level of confidence [23] [25]. This guide frames these concepts within the broader research problem of formulating a parameter estimation study, detailing methodologies, presenting quantitative comparisons, and providing practical tools for researchers and drug development professionals.
Core Definitions and Conceptual Framework
Point Estimation involves using sample data to calculate a single value (a point estimate) that serves as the best guess for an unknown population parameter, such as the population mean (μ) or proportion (p) [23] [24]. Common examples include the sample mean (x̄) estimating the population mean and the sample proportion (p̂) estimating the population proportion [23]. Its primary advantage is simplicity and direct interpretability [26]. However, a significant drawback is that it does not convey any information about its own reliability or the uncertainty associated with the estimation [23] [27].
Interval Estimation, most commonly expressed as a confidence interval, provides a range of values constructed from sample data. This range is likely to contain the true population parameter with a specified degree of confidence (e.g., 95%) [23] [25]. An interval estimate accounts for sampling variability and offers a measure of precision, making it a more robust and informative approach for scientific reporting [26] [27].
The fundamental difference lies in their treatment of uncertainty: a point estimate is a specific value, while an interval estimate explicitly quantifies uncertainty by providing a range [23].
Quantitative Comparison and Data Presentation
The following tables summarize key differences and example calculations for point and interval estimates.
Table 1: Conceptual and Practical Differences
| Aspect | Point Estimate | Interval Estimate |
|---|---|---|
| Definition | A single value estimate of a population parameter. [23] | A range of values used to estimate a population parameter. [23] |
| Precision & Uncertainty | Provides a specific value but does not reflect uncertainty or sampling variability. [23] | Provides a range that accounts for sampling variability, reflecting uncertainty. [23] |
| Confidence Level | Not applicable. | Accompanied by a confidence level (e.g., 95%) indicating the probability the interval contains the true parameter. [23] |
| Primary Use Case | Simple communication of a "best guess"; input for downstream deterministic calculations. [26] | Conveying the reliability and precision of an estimate; basis for statistical inference. [27] |
| Information Conveyed | Location. | Location and precision. |
Table 2: Common Point Estimate Calculations [23]
| Parameter | Point Estimator | Formula |
|---|---|---|
| Population Mean (μ) | Sample Mean (x̄) | x̄ = (1/n) Σ X_i |
| Population Proportion (p) | Sample Proportion (p̂) | p̂ = x / n |
| Population Variance (σ²) | Sample Variance (s²) | s² = [1/(n-1)] Σ (X_i - x̄)² |
Table 3: Common Interval Estimate (Confidence Interval) Calculations [23]
| Parameter (Assumption) | Confidence Interval Formula | Key Components |
|---|---|---|
| Mean (σ known) | x̄ ± z*(σ/√n) | z: Critical value from standard normal distribution. |
| Mean (σ unknown) | x̄ ± t*(s/√n) | t: Critical value from t-distribution with (n-1) df. |
| Proportion | p̂ ± z*√[p̂(1-p̂)/n] | z: Critical value; n: sample size. |
Table 4: Example from Drug Development Research (Illustrative Parameters) [28]
| Therapeutic Area | Phase 3 Avg. Duration (Months) | Phase 3 Avg. Patients per Trial | Notes on Estimation |
|---|---|---|---|
| Overall Average | 38.0 | 630 | Weighted averages from multiple databases (Medidata, clinicaltrials.gov, FDA DASH). [28] |
| Pain & Anesthesia | Not Specified | 1,209 | High variability in patient enrollment across therapeutic areas. [28] |
| Hematology | Not Specified | 233 | Demonstrates the range of values, underscoring the need for interval estimates. [28] |
Experimental Protocols for Estimation
Formulating a parameter estimation problem requires a structured methodology. Below are detailed protocols for key estimation approaches.
Protocol for Deriving a Point Estimate via Maximum Likelihood Estimation (MLE)
Objective: To find the parameter value that maximizes the probability (likelihood) of observing the given sample data. Procedure:
- Model Specification: Define the probability density (or mass) function for the data, f(x; θ), where θ is the unknown parameter(s). [24]
- Likelihood Function Construction: Given a random sample (X₁, X₂, ..., Xₙ), form the likelihood function L(θ) = ∏ f(xᵢ; θ), the joint probability of the sample. [24]
- Log-Likelihood: Compute the log-likelihood function, ℓ(θ) = log L(θ), to simplify differentiation. [24]
- Maximization: Take the derivative of ℓ(θ) with respect to θ, set it equal to zero: dℓ(θ)/dθ = 0. [24]
- Solution: Solve the resulting equation(s) for θ. The solution, denoted θ̂MLE, is the maximum likelihood estimate. [24] [29] Interpretation: θ̂MLE is the parameter value under which the observed data are most probable.
Protocol for Deriving a Point Estimate via Method of Moments (MOM)
Objective: To estimate parameters by equating sample moments to theoretical population moments. Procedure:
- Identify Parameters: Determine the number (k) of unknown parameters to estimate. [24]
- Express Population Moments: Calculate the first 'k' theoretical population moments (e.g., mean μ, variance σ²) as functions of the unknown parameters. [24]
- Calculate Sample Moments: Calculate the corresponding sample moments from the data. For the r-th moment: m_r = (1/n) Σ Xᵢʳ. [24]
- Equate and Solve: Set the population moment equations equal to the sample moments: μr(θ) = mr for r = 1, ..., k. Solve this system of equations for the parameters. [24] [29] Interpretation: The MOM estimate provides a simple, intuitive estimator that may be less efficient than MLE but is often easier to compute.
Protocol for Constructing a Confidence Interval for a Population Mean (σ Unknown)
Objective: To construct a range that has a 95% probability of containing the true population mean μ. Procedure:
- Collect Data: Obtain a random sample of size n, recording values X₁, X₂, ..., Xₙ.
- Calculate Sample Statistics: Compute the sample mean (x̄) and sample standard deviation (s). [23]
- Determine Critical Value: Based on the desired confidence level (e.g., 95%) and degrees of freedom (df = n-1), find the two-tailed critical t-value (t*) from the t-distribution table. [23]
- Compute Standard Error: Calculate the standard error of the mean: SE = s / √n. [23]
- Calculate Margin of Error: ME = t* × SE. [23]
- Construct Interval: The 95% confidence interval is: (x̄ - ME, x̄ + ME). [23] Interpretation: We are 95% confident that this interval captures the true population mean μ. The "95% confidence" refers to the long-run success rate of the procedure. [30]
Visualization of the Parameter Estimation Workflow
Title: Workflow for Statistical Parameter Estimation from Sample Data
Types of Intervals and Their Relationships
It is crucial to distinguish between confidence intervals, prediction intervals, and tolerance intervals, as they address different questions [30].
Title: Comparison of Confidence, Prediction, and Tolerance Intervals
- Confidence Interval (CI): Quantifies uncertainty about a population parameter (e.g., mean). It becomes narrower with increased sample size, theoretically collapsing to the true parameter value [30].
- Prediction Interval (PI): Quantifies uncertainty about a future individual observation. It is wider than a CI as it must account for both parameter uncertainty and individual data point variability. It does not converge to a single value as sample size increases [30].
- Tolerance Interval (TI): Provides a range that, with a specified confidence level (e.g., 95%), contains at least a specified proportion (e.g., 95%) of the population. It is used for quality control and setting specification limits [30].
The Scientist's Toolkit: Research Reagent Solutions
Table 5: Essential Tools for Parameter Estimation Research
| Tool / Reagent | Function in Estimation Research |
|---|---|
| Maximum Likelihood Estimation (MLE) | A fundamental algorithm for finding point estimates that maximize the probability of observed data. Critical for fitting complex models (e.g., PBPK). [24] [31] |
| Method of Moments (MOM) | A simpler, sometimes less efficient, alternative to MLE for deriving initial point estimates. [24] [29] |
| Nonlinear Least-Squares Solvers | Core algorithms (e.g., quasi-Newton, Nelder-Mead) for estimating parameters by minimizing the difference between model predictions and observed data, especially in pharmacokinetics. [31] |
| Bootstrapping Software | A resampling technique used to construct non-parametric confidence intervals, especially useful when theoretical distributions are unknown. [23] |
| Markov Chain Monte Carlo (MCMC) | A Bayesian estimation method for sampling from complex posterior distributions to obtain point estimates (e.g., posterior mean) and credible intervals (Bayesian analog of CI). [24] |
| Statistical Software (R, Python SciPy/Statsmodels) | Provides libraries for executing MLE, computing confidence intervals, and performing bootstrapping. [24] |
| PBPK/QSP Modeling Platforms | Specialized software (e.g., GastroPlus, Simcyp) that integrate parameter estimation algorithms to calibrate complex physiological models against experimental data. [31] |
Formulating the Parameter Estimation Problem within Research
A well-formulated parameter estimation problem is the backbone of quantitative research. The process involves:
- Defining the Target Parameter (θ): Clearly state the population quantity of interest (e.g., mean drug development phase duration, efficacy proportion). [28] [29]
- Selecting the Estimation Paradigm: Choose between point or interval estimation based on the research goal. For reporting final results or assessing precision, interval estimates are mandatory. For initial model fitting or inputs to deterministic models, point estimates may suffice. [26]
- Choosing an Estimator and Method: Select an appropriate statistic (e.g., sample mean, MLE) and a method for calculating it (or its interval). Considerations include bias, efficiency, and model assumptions. [24] [27]
- Assessing Estimator Properties: Evaluate the chosen estimator's unbiasedness, consistency, and efficiency within the context of the study. [24]
- Implementing and Validating: Use appropriate algorithms and software to perform the estimation. For complex models, use multiple algorithms and initial values to ensure credible results. [31]
- Contextual Interpretation: Report estimates within the research narrative. A point estimate provides a central value, but an interval estimate formally communicates the uncertainty, which is critical for risk assessment in fields like drug development [28] and for making robust scientific conclusions [26].
The Optimization Blueprint: Translating Your Problem into a Solvable Formulation
In scientific research and industrial development, the formulation of a parameter estimation problem is foundational for building predictive mathematical models. Central to this process is the systematic definition of design variables—the specific parameters and initial states whose values must be determined from experimental data to calibrate a model. In the context of Model-Based Design of Experiments (MBDoE), design variables represent the unknown quantities in a mathematical model that researchers aim to estimate with maximum precision through strategically designed experiments [15]. The careful identification of these variables is a critical first step that directly influences the reliability of the resulting model and the efficiency of the entire experimental process.
In technical domains such as chemical process engineering and pharmaceutical development, models—whether mechanistic, data-driven, or semi-empirical—serve as quantitative representations of system behavior [15]. The parameter precision achieved through estimation dictates a model's predictive power and practical utility. A well-formulated set of design variables enables researchers to focus experimental resources on obtaining the most informative data, thereby accelerating model calibration and reducing resource consumption. This guide provides a comprehensive framework for identifying and classifying these essential elements within a parameter estimation problem, with specific applications in drug development and process engineering.
Theoretical Foundations and Classification
Core Components of Design Variables
In a parameter estimation problem, design variables can be systematically categorized into two primary groups:
- Model Parameters: These are intrinsic properties of the system that remain constant under the defined experimental conditions but are unknown a priori. Examples include kinetic rate constants in a reaction network, thermodynamic coefficients, material properties, and affinity constants in pharmacological models [15] [10].
- Initial States: These variables define the starting conditions of the system at the beginning of an experiment or observational period. Unlike parameters, initial states are often set by the experimenter but may also be unknown and require estimation for model initialization. Examples include initial reactant concentrations in a batch reactor and baseline biomarker levels in a physiological system [15].
The process of identifying these variables requires a deep understanding of the system's underlying mechanisms. The relationship between a model's output and its design variables is typically expressed as:
y = f(t, θ, x₀, u)
Where:
- y is the model output or response variable.
- f is the mathematical representation of the system.
- t is time (or an independent variable).
- θ is the vector of model parameters to be estimated.
- x₀ is the vector of initial states.
- u is the vector of controllable input variables or experimental design factors.
The MBDoE Framework for Parameter Precision
Model-Based Design of Experiments (MBDoE) is a structured methodology for designing experiments that maximize the information content of the collected data for the specific purpose of parameter estimation [15]. Within this framework, the quality of parameter estimates is often quantified using the Fisher Information Matrix (FIM). The FIM is inversely related to the variance-covariance matrix of the parameter estimates, and its maximization is a central objective in MBDoE. The FIM (M) for a dynamic system is calculated as:
M = ∑ᵢ (∂y/∂θ)ᵀ Q⁻¹ (∂y/∂θ)
Where:
- The summation is over all experimental samples.
- ∂y/∂θ is the sensitivity matrix of the model outputs with respect to the parameters.
- Q is the variance-covariance matrix of the measurement errors.
The primary goal is to design experiments that maximize a scalar function of the FIM (e.g., its determinant, known as D-optimality), which directly leads to minimized confidence regions for the estimated parameters, θ [15].
Methodologies for Identifying Design Variables
A systematic, multi-stage approach is required to reliably identify the parameters and initial states that constitute the design variables for estimation. The following workflow outlines this process, from initial model conceptualization to the final selection of variables for the experimental design.
A Systematic Workflow for Variable Identification
The following diagram illustrates the logical sequence and iterative nature of the identification process.
Detailed Experimental Protocols
For each key step in the workflow, a specific methodological approach is required.
Protocol for Preliminary Sensitivity Analysis (Step 2)
- Objective: To rank candidate parameters based on their influence on model outputs.
- Procedure:
- Define a nominal parameter vector (θ₀) and a plausible range for each parameter based on literature or expert knowledge.
- Simulate the model output, y(t, θ₀).
- For each parameter θᵢ, compute the local sensitivity coefficient: Sᵢ(t) = ∂y(t)/∂θᵢ. This is often done via finite differences or by solving associated sensitivity equations.
- Aggregate sensitivity measures (e.g., the L₂-norm of Sᵢ(t) over the time course) to rank parameters.
- Output: A ranked list of parameters, prioritizing those with the highest sensitivity for inclusion as design variables.
Protocol for Assessing Practical Identifiability (Step 5)
- Objective: To determine if the available experimental data is sufficient to uniquely estimate the selected parameters.
- Procedure:
- Using initial experimental data, compute the FIM.
- Check the condition number of the FIM. A very high condition number indicates potential collinearity between parameters (non-identifiability).
- Compute the profile likelihood for each parameter. This involves varying one parameter while re-optimizing all others and plotting the resulting cost function value.
- A uniquely identifiable parameter will show a well-defined, concave minimum in its profile likelihood. A flat profile indicates non-identifiability.
- Output: A diagnosis of which parameters are practically identifiable, guiding model simplification or experimental redesign.
Quantitative Data and Application in Drug Development
Quantitative Analysis of Design Variable Impact
The table below summarizes key quantitative findings from recent research, highlighting the performance of advanced methodologies in managing design variables for parameter estimation.
Table 1: Quantitative Performance of Advanced Frameworks in Parameter Estimation and Variable Optimization
| Framework/Method | Primary Application | Key Performance Metric | Reported Value | Implication for Design Variables |
|---|---|---|---|---|
| optSAE + HSAPSO [32] | Drug classification & target identification | Classification Accuracy | 95.52% | Demonstrates high precision in identifying relevant biological parameters from complex data. |
| Computational Efficiency | 0.010 s/sample | Enables rapid, iterative testing of different variable sets and model structures. | ||
| Stability (Variability) | ± 0.003 | Indifies robust parameter estimates with low sensitivity to noise, confirming good variable selection. | ||
| MBDoE Techniques [15] | Chemical process model calibration | Parameter Precision | Up to 40% improvement vs. OFAT | Systematically designed experiments around key variables drastically reduce confidence intervals of estimates. |
| HSAPSO-SAE [32] | Pharmaceutical informatics | Hyperparameter Optimization | Adaptive tuning of SAE parameters | Validates the role of meta-optimization (optimizing the optimizer's parameters) for handling complex variable spaces. |
| Fit-for-Purpose MIDD [10] | Model-Informed Drug Development | Contextual Alignment | Alignment with QOI/COU | Ensures the selected design variables are directly relevant to the specific drug development question. |
Application in Pharmaceutical Development
In Model-Informed Drug Development (MIDD), the "fit-for-purpose" principle dictates that the selection of design variables must be closely aligned with the Key Questions of Interest (QOI) and Context of Use (COU) at each stage [10]. The following table maps common design variables to specific drug development activities, illustrating this alignment.
Table 2: Design Variables and Their Roles in Stages of Drug Development
| Drug Development Stage | Common MIDD Tool(s) | Typical Design Variables (Parameters & Initial States) for Estimation | Purpose of Estimation |
|---|---|---|---|
| Discovery | QSAR [10] | - IC₅₀- Binding affinity constants- Physicochemical properties (logP, pKa) | Prioritize lead compounds based on predicted activity and properties. |
| Preclinical | PBPK [10] | - Tissue-to-plasma partition coefficients- Clearance rates- Initial organ concentrations | Predict human pharmacokinetics and safe starting dose for First-in-Human (FIH) trials. |
| Clinical (Phase I) | Population PK (PPK) [10] | - Volume of distribution (Vd)- Clearance (CL)- Inter-individual variability (IIV) on parameters | Characterize drug exposure and its variability in a human population. |
| Clinical (Phase II/III) | Exposure-Response (E-R) [10] | - E₀ (Baseline effect)- Eₘₐₓ (Maximal effect)- EC₅₀ (Exposure for 50% effect) | Quantify the relationship between drug exposure and efficacy/safety outcomes to inform dosing. |
The hierarchically self-adaptive particle swarm optimization (HSAPSO) algorithm, cited in Table 1, exemplifies a modern approach to handling complex variable estimation. It dynamically adjusts its own parameters during optimization, leading to more efficient and reliable convergence on the optimal values for the primary model parameters (θ) [32].
The Scientist's Toolkit: Essential Research Reagents and Materials
The successful execution of experiments for parameter estimation relies on a suite of computational and experimental tools. The following table details key solutions and their functions.
Table 3: Key Research Reagent Solutions for Parameter Estimation Experiments
| Category / Item | Specific Examples | Function in Variable Identification & Estimation |
|---|---|---|
| Modeling & Simulation Software | MATLAB, SimBiology, R, Python (SciPy, PyMC3), NONMEM, Monolix | Provides the computational environment to implement mathematical models, perform simulations, and execute parameter estimation algorithms. |
| Sensitivity Analysis Tools | Sobol' method (for global SA), Morris method, software-integrated solvers | Quantifies the influence of each candidate parameter on model outputs, guiding the selection of design variables for estimation. |
| Optimization Algorithms | Particle Swarm Optimization (PSO) [32], Maximum Likelihood Estimation (MLE), Bayesian Estimation | The core computational engine for finding the parameter values that minimize the difference between model predictions and experimental data. |
| Model-Based DoE Platforms | gPROMS FormulatedProducts, CADRE | Specialized software to design optimal experiments that maximize information gain for the specific set of design variables. |
| Data Management & Curation | Electronic Lab Notebooks (ELNs), SQL databases, FAIR data principles | Ensures the quality, traceability, and accessibility of experimental data used for parameter estimation, which is critical for reliable results. |
| High-Throughput Screening Assays | Biochemical activity assays, ADME (Absorption, Distribution, Metabolism, Excretion) profiling | Generates rich, quantitative datasets from which parameters like IC₅₀, clearance, and permeability can be estimated. |
The precise definition of design variables—the parameters and initial states to be estimated—is the cornerstone of formulating a robust parameter estimation problem. This process is not a one-time event but an iterative cycle of model hypothesizing, variable sensitivity testing, and experimental design refinement. As demonstrated by advanced applications in drug development, a disciplined and "fit-for-purpose" approach to variable selection, supported by methodologies like MBDoE and modern optimization algorithms, is critical for building models with high predictive power. This, in turn, accelerates scientific discovery and de-risks development processes in high-stakes fields like pharmaceuticals and chemical engineering.
In scientific and engineering disciplines, particularly in pharmaceutical development and systems biology, the process of parameter estimation is fundamental for building accurate mathematical models from observed data. This process involves calibrating model parameters so that the model's output closely matches experimental measurements. The core of this calibration lies in the objective function (also known as a cost or loss function), which quantifies the discrepancy between model predictions and observed data [33] [34]. The formulation of this function directly influences which parameter values will be identified as optimal, making its selection a critical step in model development.
Parameter estimation is fundamentally framed as an optimization problem where the solution is the set of parameter values that minimizes this discrepancy [33]. This optimization problem consists of several key components: the design variables (parameters to be estimated), the objective function that measures model-data discrepancy, and optional bounds or constraints on parameter values based on prior knowledge [33]. Within this framework, the choice of objective function determines how "goodness-of-fit" is quantified, with different measures having distinct statistical properties, computational characteristics, and sensitivities to various data features.
Fundamental Measures of Fit
Sum of Squared Errors (SSE)
The Sum of Squared Errors (SSE) is one of the most prevalent objective functions in scientific modeling. It is defined as the sum of the squared differences between observed values and model predictions [33] [34]. For a dataset with N observations, the SSE formulation is:
[ \text{SSE} = \sum{i=1}^{N} (y{i} - f(xi, \theta))^2 = \sum{i=1}^{N} e_i^2 ]
where (yi) represents the actual observed value, (f(xi, \theta)) is the model prediction given inputs (xi) and parameters (\theta), and (ei) is the error residual for the (i)-th data point [33] [34].
The SSE loss function has an intuitive geometric interpretation – it represents the total area of squares constructed on the errors between data points and the model curve [34]. In this geometric context, finding the parameter values that minimize the SSE is equivalent to finding the model that minimizes the total area of these squares.
A common variant is the Mean Squared Error (MSE), which divides the SSE by the number of observations [35]. While MSE and SSE share the same minimizer (the same parameter values minimize both functions), MSE offers practical advantages in optimization algorithms like gradient descent by maintaining smaller gradient values, which often leads to more stable and efficient convergence [35].
Sum of Absolute Errors (SAE)
The Sum of Absolute Errors (SAE), also known as the Sum of Absolute Deviations (SAD), provides an alternative approach to quantifying model-data discrepancy [33] [36]. The SAE is defined as:
[ \text{SAE} = \sum{i=1}^{N} |y{i} - f(x_i, \theta)| ]
Unlike SSE, which squares the errors, SAE uses the absolute value of each error term [33] [36]. This fundamental difference in mathematical formulation leads to distinct properties in how the two measures handle variations between predictions and observations.
SAE is particularly valued for its robustness to outliers in the data [37]. Because SAE does not square the errors, it gives less weight to large residuals compared to SSE, making the resulting parameter estimates less sensitive to extreme or anomalous data points [37]. This characteristic makes SAE preferable in situations where the data may contain significant measurement errors or when the underlying error distribution has heavy tails.
Maximum Likelihood Estimation (MLE)
Maximum Likelihood Estimation (MLE) takes a fundamentally different approach by framing parameter estimation as a statistical inference problem [38] [39]. Rather than directly minimizing a measure of distance between predictions and observations, MLE seeks the parameter values that make the observed data most probable under the assumed statistical model [39].
The core concept in MLE is the likelihood function. For a set of observed data points (x1, x2, \ldots, x_n), and parameters (\theta), the likelihood function (L(\theta)) is defined as the joint probability (or probability density) of observing the data given the parameters [38] [39]:
[ L(\theta) = P(X1=x1, X2=x2, \ldots, Xn=xn; \theta) ]
for discrete random variables, or
[ L(\theta) = f(x1, x2, \ldots, x_n; \theta) ]
for continuous random variables, where (f) is the joint probability density function [38].
The maximum likelihood estimate (\hat{\theta}) is the parameter value that maximizes this likelihood function [39]:
[ \hat{\theta} = \underset{\theta}{\operatorname{arg\,max}} \, L(\theta) ]
In practice, it is often convenient to work with the log-likelihood (\ell(\theta) = \ln L(\theta)), as products in the likelihood become sums in the log-likelihood, which simplifies differentiation and computation [39].
Table 1: Comparison of Fundamental Objective Functions
| Measure | Mathematical Formulation | Key Properties | Common Applications |
|---|---|---|---|
| Sum of Squared Errors (SSE) | (\sum{i=1}^{N} (yi - f(x_i, \theta))^2) | Differentiable, sensitive to outliers, maximum likelihood for normal errors | Linear regression, nonlinear least squares, model calibration |
| Sum of Absolute Errors (SAE) | (\sum{i=1}^{N} |yi - f(x_i, \theta)|) | Robust to outliers, non-differentiable at zero | Robust regression, applications with outlier contamination |
| Maximum Likelihood (MLE) | (\prod{i=1}^{N} P(yi | xi, \theta)) or (\prod{i=1}^{N} f(yi | xi, \theta)) | Statistically efficient, requires error distribution specification | Statistical modeling, parametric inference, generalized linear models |
Theoretical Foundations and Relationships
Statistical Interpretations
The choice between SSE and SAE objective functions has profound statistical implications that extend beyond mere optimization. When errors are independent and identically distributed according to a normal distribution, minimizing the SSE yields parameter estimates that are also maximum likelihood estimators [39] [37]. This connection provides a solid statistical foundation for the SSE objective function under the assumption of normally distributed errors.
The relationship between SSE and MLE becomes clear when we consider the normal distribution explicitly. For a normal error distribution with constant variance, the log-likelihood function is proportional to the negative SSE, which means maximizing the likelihood is equivalent to minimizing the sum of squares [39]. This important relationship explains why SSE has been so widely adopted in statistical modeling – it represents the optimal estimation method when the Gaussian assumption holds.
For SAE, a similar connection exists to the Laplace distribution. If errors follow a Laplace (double exponential) distribution, then maximizing the likelihood is equivalent to minimizing the sum of absolute errors [37]. This provides a statistical justification for SAE when errors have heavier tails than the normal distribution.
Geometric and Computational Considerations
From a geometric perspective, SSE and SAE objective functions lead to different solution characteristics in optimization problems. The SSE function is differentiable everywhere, which enables the use of efficient gradient-based optimization methods [33] [34]. The resulting optimization landscape is generally smooth, often with a single minimum for well-behaved models.
In contrast, the SAE objective function is not differentiable at points where residuals equal zero, which can present challenges for some optimization algorithms [37]. However, SAE has the advantage of being more robust to outliers because it does not square the errors, thereby giving less weight to extreme deviations compared to SSE [37].
The following diagram illustrates the workflow for selecting an appropriate objective function based on data characteristics and modeling goals:
Implementation and Optimization Frameworks
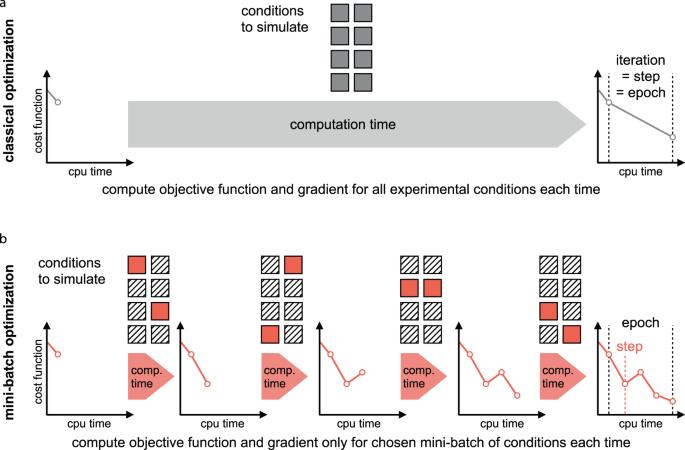
Optimization Methods and Problem Formulation
The choice of objective function directly influences the selection of appropriate optimization algorithms. Different optimization methods are designed to handle specific types of objective functions and constraints [33]. The table below summarizes common optimization methods and their compatibility with different objective function formulations:
Table 2: Optimization Methods for Different Objective Functions
| Optimization Method | Compatible Objective Functions | Problem Formulation | Key Characteristics |
|---|---|---|---|
| Nonlinear Least Squares (lsqnonlin) | SSE, Residuals | (\min\limitsx |F(x)|2^2) subject to bounds | Efficient for SSE minimization, requires fixed time base [33] |
| Gradient Descent (fmincon) | SSE, SAE, Custom functions | (\min\limits_x F(x)) subject to bounds and constraints | General-purpose, handles custom cost functions and constraints [33] |
| Simplex Search (fminsearch) | SSE, SAE | (\min\limits_x F(x)) | Derivative-free, handles non-differentiable functions [33] |
| Pattern Search | SSE, SAE | (\min\limits_x F(x)) subject to bounds | Direct search method, handles non-smooth functions [33] |
The optimization problem can be formulated in different ways depending on the modeling objectives. A minimization problem focuses solely on minimizing the objective function (F(x)). A mixed minimization and feasibility problem minimizes (F(x)) while satisfying specified bounds and constraints (C(x)). A feasibility problem focuses only on finding parameter values that satisfy constraints, which is less common in parameter estimation [33].
Practical Implementation Considerations
In practical implementation, several factors influence the success of parameter estimation using these objective functions. The time base for evaluation must be carefully considered, as measured and simulated signals may have different time bases [33]. The software typically evaluates the objective function only for the time interval common to both signals, using interpolation when necessary.
Parameter bounds based on prior knowledge of the system can significantly improve estimation results [33]. For example, in pharmaceutical applications, parameters representing physiological quantities (such as organ volumes or blood flow rates) should be constrained to biologically plausible ranges. These bounds are expressed as:
[ \underline{x} \leq x \leq \overline{x} ]
where (\underline{x}) and (\overline{x}) represent lower and upper bounds for the design variables [33].
Additionally, multi-parameter constraints can encode relationships between parameters. For instance, in a friction model, a constraint might specify that the static friction coefficient must be greater than or equal to the dynamic friction coefficient [33]. Such constraints are expressed as (C(x) \leq 0) and can be incorporated into the optimization problem.
Experimental Protocols and Applications
Protocol for Comparative Assessment of Objective Functions
To ensure robust parameter estimation in practice, researchers should implement a systematic protocol for comparing different objective functions and optimization methods:
Problem Formulation: Clearly define the model structure, parameters to be estimated, and available experimental data. Establish biologically or physically plausible parameter bounds based on prior knowledge [33] [31].
Multiple Algorithm Implementation: Implement parameter estimation using at least two different optimization algorithms (e.g., gradient-based and direct search methods) to mitigate algorithm-specific biases [31].
Objective Function Comparison: Conduct estimation using both SSE and SAE objective functions to assess sensitivity to outliers [37].
Initial Value Sensitivity Analysis: Perform estimation from multiple different initial parameter values to check for local minima and assess solution stability [31].
Model Validation: Compare the predictive performance of the resulting parameter estimates on a separate validation dataset not used during estimation.
Uncertainty Quantification: Where possible, compute confidence intervals for parameter estimates using appropriate statistical techniques (e.g., profile likelihood, bootstrap methods).
This comprehensive approach helps ensure that the final parameter estimates are credible and not unduly influenced by the specific choice of objective function or optimization algorithm [31].
Application in Pharmaceutical Sciences
In Physiologically-Based Pharmacokinetic (PBPK) and Quantitative Systems Pharmacology (QSP) modeling, parameter estimation is typically performed using the nonlinear least-squares method, which minimizes the SSE between model predictions and observed data [31]. The following diagram illustrates the integrated parameter estimation workflow in pharmaceutical modeling:
Comparative studies have demonstrated that parameter estimation results can be significantly influenced by the choice of initial values and that the best-performing algorithm often depends on factors such as model structure and the specific parameters being estimated [31]. Therefore, employing multiple estimation algorithms under different conditions is recommended to obtain credible parameter estimates [31].
Research Reagent Solutions
Table 3: Essential Computational Tools for Parameter Estimation
| Tool Category | Representative Examples | Function in Parameter Estimation |
|---|---|---|
| Optimization Software | MATLAB Optimization Toolbox, SciPy Optimize | Provides algorithms (lsqnonlin, fmincon, fminsearch) for minimizing objective functions [33] |
| Modeling Environments | Simulink, Berkeley Madonna, COPASI | Enable construction of complex models and provide built-in parameter estimation capabilities |
| Statistical Packages | R, Python Statsmodels, SAS | Offer maximum likelihood estimation and specialized regression techniques |
| Custom Code Templates | SSE/SAE/MLE functions, Gradient descent | Implement specific objective functions and optimization workflows [37] |
The formulation of the objective function represents a critical decision point in the parameter estimation process, with significant implications for the resulting model parameters and their biological or physical interpretation. The Sum of Squared Errors offers optimal properties when errors are normally distributed and enables efficient optimization through differentiable methods. The Sum of Absolute Errors provides robustness to outliers and is preferable when the error distribution has heavy tails. Maximum Likelihood Estimation offers a principled statistical framework that can incorporate specific assumptions about error distributions.
In practice, the selection of an objective function should be guided by the characteristics of the experimental data, the purpose of the modeling effort, and computational considerations. A comprehensive approach that tests multiple objective functions and optimization algorithms provides the most reliable path to credible parameter estimates, particularly for complex models in pharmaceutical research and systems biology. The integration of these objective functions within a rigorous model development workflow ensures that mathematical models not only fit historical data but also possess predictive capability for novel experimental conditions.
Parameter estimation is a fundamental process in building mathematical models for scientific and engineering applications, ranging from drug development to chemical process control. An accurate dynamic model is required to achieve good monitoring, online optimization, and control performance. However, most process models and measurements are corrupted by noise and modeling inaccuracies, creating the need for robust estimation of both states and parameters [40]. Often, the state and parameter estimation problems are solved simultaneously by augmenting parameters to the states, creating a nonlinear filtering problem for most systems [40].
The incorporation of bounds and constraints transforms naive parameter estimation into a scientifically rigorous procedure that incorporates prior knowledge and physical realism. In practice, constraints on parameter values can often be generated from fundamental principles, experimental evidence, or physical limitations. For example, the surface area of a reactor must be positive, and rate constants typically fall within biologically plausible ranges [40] [41]. The strategic incorporation of these constraints significantly improves estimation performance, enhances model identifiability, and ensures that estimated parameters maintain physical meaning [40] [41].
This technical guide examines the formulation, implementation, and application of constrained parameter estimation methods within a comprehensive research framework. By systematically integrating prior knowledge through mathematical constraints, researchers can develop more credible, predictive models capable of supporting critical decisions in drug development, chemical engineering, and related fields.
Theoretical Foundations of Constrained Parameter Estimation
Problem Formulation
Constrained parameter estimation extends the traditional estimation problem by incorporating known relationships and limitations into the mathematical framework. The general continuous-discrete nonlinear stochastic process model can be represented as:
Process Model: ẋ = f(x, u, θ) + w
Measurement Model: y_k = h(x_k) + v_k
where x, x_k ∈ R^n denote the vector of states, u ∈ R^q denotes the vector of known manipulated variables, and y_k ∈ R^m denotes the vector of available measurements. The function f:R^n → R^n represents the state function with parameters θ ∈ R^p, while h:R^n → R^m is the measurement function. The terms w ∈ R^n and v_k ∈ R^m represent process and measurement noise respectively, with independent distributions w ~ N(0, Q) and v_k ~ N(0, R), where Q and R are covariance matrices [40].
Parameters are commonly augmented to the states, leading to the simultaneous estimation of states and parameters. This dual estimation problem represents a nonlinear filtering problem for most chemical and biological processes [40].
Types of Constraints in Scientific Modeling
Inequality constraints on parameters typically take the form:
d_L ≤ c(θ_1, θ_2, ..., θ_p) ≤ d_U
where c(·) is a function describing the parameter relationship, and d_L and d_U indicate the lower and upper bounds of the inequality constraints [40]. These constraints may be derived from various sources of prior knowledge:
- Physical Reality Constraints: Parameters must remain within physically plausible ranges (e.g., positive rate constants, bounded concentrations) [41].
- Theoretical Domain Constraints: Values constrained by theoretical limitations (e.g., fractions between 0 and 1, probabilities summing to 1).
- Experimental Observation Constraints: Ranges consistent with previous experimental measurements.
- Stability and Performance Constraints: Limitations that ensure model stability or desired performance characteristics.
Table 1: Classification of Parameter Constraints in Scientific Models
| Constraint Type | Mathematical Form | Typical Application | Implementation Consideration |
|---|---|---|---|
| Simple Bounds | θ_L ≤ θ ≤ θ_U |
Physically plausible parameter ranges | Straightforward to implement; direct application in optimization |
| Linear Inequality | Aθ ≤ b |
Mass balance constraints, resource limitations | Requires careful construction of constraint matrices |
| Nonlinear Inequality | g(θ) ≤ 0 |
Thermodynamic relationships, complex biological limits | Computational challenges; potential for local minima |
| Sparsity Constraints | ‖Wθ‖_0 ≤ K |
Tissue-dependent parameters in MR mapping [42] | Requires specialized algorithms; combinatorial complexity |
Benefits of Constrained Estimation
The strategic incorporation of constraints provides multiple advantages throughout the modeling lifecycle:
- Improved Identifiability: Constraints resolve parameter identifiability issues by restricting the feasible solution space, particularly important in complex physiologically-based pharmacokinetic (PBPK) models with many correlated parameters [41].
- Enhanced Numerical Stability: Well-formulated constraints stabilize optimization algorithms and prevent divergence to physically meaningless solutions.
- Accelerated Convergence: By eliminating infeasible regions of parameter space, constraints reduce the search domain and speed up estimation procedures [40].
- Increased Predictive Accuracy: Constraints incorporating genuine prior knowledge improve model performance in extrapolation scenarios, essential for predicting behavior outside studied conditions [41].
Methodological Approaches for Constrained Estimation
Framework for Constraint Implementation
A robust framework for inequality constrained parameter estimation employs a two-stage approach: first solving an unconstrained estimation problem, followed by a constrained optimization step [40]. This methodology applies to recursive estimators such as the unscented Kalman filter (UKF) and the ensemble Kalman filter (EnKF), as well as moving horizon estimation (MHE).
The mathematical formulation for this framework can be represented as:
Stage 1 - Unconstrained Estimation:
{ρ̂, θ̂} = argmin Σ ‖d_m - F_mΨ_mΦ_m(θ)ρ‖₂²
Stage 2 - Constrained Refinement:
{ρ̂_c, θ̂_c} = argmin Σ ‖d_m - F_mΨ_mΦ_m(θ)ρ‖₂² subject to constraints on θ and ρ
This approach provides fast recovery of state and parameter estimates from inaccurate initial guesses, leading to better estimation and control performance [40].
Construction of Inequality Constraints from Data
Inequality constraints can be systematically derived from routine steady-state operating data, even when such data is corrupted with moderate noise. The methodology involves:
- Data Collection: Gather steady-state operational data under varying conditions.
- Parameter Relationship Identification: Analyze correlations and fundamental relationships between parameters.
- Constraint Formulation: Transform identified relationships into mathematical inequalities with appropriate bounds.
- Validation: Verify that constraints are consistent with physical principles and additional experimental data.
For example, in chemical process modeling, steady-state gain relationships between manipulated and controlled variables can be transformed into parameter constraints that improve estimation performance [40].
Algorithms for Constrained Parameter Estimation
Various computational algorithms have been developed specifically for constrained estimation problems:
- Constrained Ensemble Kalman Filter (C-EnKF): Extends the EnKF framework using constrained Monte Carlo samples and probability density function truncation for nonlinear estimation [40].
- Constrained Particle Filtering (C-PF): Incorporates constraints through specialized sampling techniques [40].
- Recursive Nonlinear Dynamic Data Reconciliation (RNDDR): Takes constraints into consideration with nonlinear state and covariance propagation based on extended Kalman filter algorithms [40].
- Unscented Recursive Nonlinear Dynamic Data Reconciliation (URNDDR): Combines UKF with RNDDR for more accurate and efficient nonlinear constrained estimation [40].
Table 2: Performance Comparison of Parameter Estimation Algorithms
| Algorithm | Strengths | Limitations | Best-Suited Applications |
|---|---|---|---|
| Quasi-Newton Method | Fast convergence; efficient for medium-scale problems | May converge to local minima; requires good initial estimates | Models with smooth objective functions and available gradients |
| Nelder-Mead Method | Derivative-free; robust to noise | Slow convergence; inefficient for high-dimensional problems | Models where gradient calculation is difficult or expensive |
| Genetic Algorithm | Global search; handles non-convex problems | Computationally intensive; parameter tuning sensitive | Complex models with multiple local minima [31] |
| Particle Swarm Optimization | Global optimization; parallelizable | May require many function evaluations; convergence not guaranteed | Black-box models where gradient information is unavailable [31] |
| Cluster Gauss-Newton Method | Handles ill-posed problems; multiple solutions | Implementation complexity; computational cost | Problems with non-unique solutions or high parameter uncertainty [31] |
Experimental Protocols and Implementation
Protocol 1: Two-Stage Constrained Estimation for Chemical Processes
Objective: Estimate parameters in nonlinear chemical process models with inequality constraints derived from steady-state operating data.
Materials and Methods:
- Process Data Collection: Acquire steady-state operating data across multiple operating conditions.
- Constraint Development: Calculate steady-state gains and relationships between variables, transforming them into parameter inequality constraints.
- Unconstrained Estimation: Initialize parameters and perform unconstrained estimation using UKF or EnKF to obtain preliminary estimates.
- Constrained Refinement: Solve the constrained optimization problem using the unconstrained estimates as initialization.
- Validation: Compare constrained and unconstrained estimates against holdout data to verify improved performance.
Key Considerations:
- The choice between UKF and EnKF depends on the problem dimension and nonlinearity characteristics.
- Constraint implementation methods should avoid inconsistency between state and parameter estimates [40].
- Computational efficiency must be balanced against estimation accuracy requirements.
Protocol 2: Sparsity-Constrained Parameter Mapping in Magnetic Resonance
Objective: Directly estimate parameter maps from undersampled k-space data with sparsity constraints on model parameters.
Materials and Methods:
- Data Acquisition: Collect undersampled k-space data using appropriate MR sequences.
- Signal Modeling: Formulate the explicit signal model relating parameters to measurements.
- Sparsity Constraints: Impose sparsity constraints on parameter maps using appropriate transforms (e.g., wavelet transforms).
- Constrained Optimization: Solve the resulting constrained maximum likelihood estimation problem using greedy-pursuit or other specialized algorithms [42].
- Performance Benchmarking: Compare results against estimation-theoretic bounds to validate performance.
Key Considerations:
- Sparsity constraints leverage the tissue-dependent nature of parameters, as the number of tissue types is typically much smaller than the number of voxels [42].
- Both the parameter map and spin density distribution can be constrained simultaneously.
- Orthonormal sparsifying transforms simplify the optimization problem.
Protocol 3: Middle-Out Approach for PBPK Model Calibration
Objective: Estimate uncertain parameters in complex PBPK models while maintaining physiological plausibility.
Materials and Methods:
- Parameter Classification: Separate parameters into well-identified (fixed), uncertain (estimated), and non-identifiable (constrained) categories.
- Sensitivity Analysis: Perform global sensitivity analysis to identify parameters with significant influence on model outputs.
- Structural Identifiability Analysis: Determine which parameters are theoretically identifiable from available data.
- Constrained Optimization: Estimate sensitive, identifiable parameters while applying constraints to maintain physiological plausibility [41].
- Uncertainty Quantification: Characterize parameter uncertainty and correlation structure.
Key Considerations:
- Address high parameter correlations through reparameterization or composite variables [41].
- Validate model performance against data not used in estimation, particularly in drug-drug interaction scenarios or special populations.
- Consider Bayesian approaches to incorporate prior knowledge probabilistically rather than through hard constraints.
Visualization of Constrained Estimation Workflows
Workflow for Two-Stage Constrained Parameter Estimation
Algorithm Selection Decision Framework
PBPK Model Parameter Identification Workflow
Research Reagent Solutions for Parameter Estimation
Table 3: Essential Computational Tools for Constrained Parameter Estimation
| Tool/Category | Function | Application Context | Implementation Considerations |
|---|---|---|---|
| Nonlinear Optimization Libraries (IPOPT, NLopt) | Solve constrained nonlinear optimization problems | General parameter estimation with constraints | Requires gradient information; appropriate for medium to large-scale problems |
| Kalman Filter Variants (UKF, EnKF) | Sequential state and parameter estimation | Real-time applications; chemical process control [40] | UKF better for moderate nonlinearities; EnKF suitable for high-dimensional problems |
| Global Optimization Algorithms (Genetic Algorithms, Particle Swarm) | Find global optimum in non-convex problems | Complex models with multiple local minima [31] | Computationally intensive; requires parameter tuning |
| Sensitivity Analysis Tools (Sobol, Morris Methods) | Identify influential parameters | PBPK model development; experimental design [41] | Helps prioritize parameters for estimation versus constraint |
| Identifiability Analysis Software (DAISY, GenSSI) | Assess theoretical parameter identifiability | PBPK models; systems biology [41] | Should be performed before estimation attempts |
| Bayesian Inference Frameworks (Stan, PyMC) | Probabilistic parameter estimation | Incorporation of uncertainty and prior distributions | Computationally demanding but provides full uncertainty quantification |
The strategic incorporation of bounds and constraints represents a fundamental advancement in parameter estimation methodology, transforming it from a purely mathematical exercise to a scientifically rigorous procedure that respects physical reality and prior knowledge. By systematically integrating constraints derived from first principles, experimental evidence, and domain expertise, researchers can develop more credible, predictive models capable of supporting critical decisions in drug development, chemical process optimization, and medical imaging.
The two-stage framework—initial unconstrained estimation followed by constrained refinement—provides a robust methodology applicable across diverse domains from chemical process control to magnetic resonance parameter mapping. Furthermore, the careful selection of estimation algorithms based on problem characteristics, coupled with comprehensive identifiability and sensitivity analyses, ensures that constrained parameter estimation delivers physiologically plausible results with enhanced predictive capability.
As model complexity continues to grow in fields such as quantitative systems pharmacology and physiological modeling, the disciplined application of constrained estimation methods will remain essential for developing trustworthy models that can successfully extrapolate beyond directly observed conditions—the ultimate test of model utility in scientific research and practical applications.
Parameter estimation is a fundamental inverse problem in computational science, involving the prediction of model parameters from observed data [3]. The selection of an appropriate optimization algorithm is crucial for obtaining reliable, accurate, and computationally efficient estimates. This guide provides an in-depth technical comparison of three prominent optimization methods—lsqnonlin, fmincon, and Particle Swarm Optimization (PSO)—within the context of formulating parameter estimation problems for research applications. We examine their mathematical foundations, implementation requirements, performance characteristics, and suitability for different problem classes, with particular attention to challenges in biological and pharmacological modeling where data may be sparse and models highly nonlinear [3].
Mathematical Formulations and Problem Scope
lsqnonlin: Nonlinear Least-Squares Specialist
The lsqnonlin solver addresses nonlinear least-squares problems of the form:
[
\min{x} \|f(x)\|2^2 = \min{x} \left(f1(x)^2 + f2(x)^2 + \dots + fn(x)^2\right)
]
where (f(x)) is a vector-valued function that returns a vector of residuals, not the sum of squares [43] [44]. This formulation makes it ideally suited for data-fitting applications where the goal is to minimize the difference between model predictions and experimental observations. The solver can handle bound constraints of the form (lb \leq x \leq ub), along with linear and nonlinear constraints [43].
fmincon: General-Purpose Constrained Minimization
The fmincon solver solves more general nonlinear programming problems:
[
\min_{x} f(x) \quad \text{such that} \quad \begin{cases} c(x) \leq 0 \ ceq(x) = 0 \ A \cdot x \leq b \ Aeq \cdot x = beq \ lb \leq x \leq ub \end{cases}
]
where (f(x)) is a scalar objective function, and constraints can include nonlinear inequalities (c(x)), nonlinear equalities (ceq(x)), linear inequalities, linear equalities, and bound constraints [45]. This general formulation allows fmincon to address a broader class of problems beyond least-squares minimization.
Particle Swarm Optimization: Gradient-Free Metaheuristic
Particle Swarm Optimization is a population-based metaheuristic that optimizes a problem by iteratively improving candidate solutions with regard to a given measure of quality. It solves a problem by having a population (swarm) of candidate solutions (particles), and moving these particles around in the search-space according to simple mathematical formulae over the particle's position and velocity [46]. Each particle's movement is influenced by its local best-known position and is also guided toward the best-known positions in the search-space, which are updated as better positions are found by other particles [46]. Unlike gradient-based methods, PSO does not require the problem to be differentiable, making it suitable for non-smooth, noisy, or discontinuous objective functions.
Algorithm Selection and Performance Characteristics
Computational Efficiency and Problem Structure
The choice between lsqnonlin and fmincon for solving constrained nonlinear least-squares problems involves important efficiency considerations. When applied to the same least-squares problem, lsqnonlin typically requires fewer function evaluations and iterations than fmincon [47]. This performance advantage stems from fundamental differences in how these algorithms access problem information: lsqnonlin works with the entire vector of residuals (F(x)), providing the algorithm with more detailed information about the objective function structure, while fmincon only has access to the scalar value of the sum of squares (\|F(x)\|^2) [47].
Table 1: Comparative Performance of lsqnonlin and fmincon on Constrained Least-Squares Problems
| Performance Metric | lsqnonlin | fmincon | Notes |
|---|---|---|---|
| Number of iterations | Lower | Approximately double | Difference increases with problem size N [47] |
| Function count | Lower | Higher | Consistent across derivative estimation methods [47] |
| Residual values | Equivalent | Equivalent | Results independent of solver choice [47] |
| Derivative estimation | More efficient with automatic differentiation | Shows similar improvements | Finite differences significantly increase computation [47] |
This performance difference becomes increasingly pronounced as problem dimensionality grows. Empirical evidence demonstrates that for problems of size (N) (where the number of variables is (2N)), the number of iterations for fmincon is more than double that of lsqnonlin and increases approximately linearly with (N) [47].
Constraint Handling Capabilities
Each algorithm offers different capabilities for handling constraints:
- lsqnonlin: Primarily handles bound constraints ((lb \leq x \leq ub)) but can also address linear and nonlinear constraints through additional input arguments [43].
- fmincon: Provides comprehensive constraint handling for linear inequalities ((A \cdot x \leq b)), linear equalities ((Aeq \cdot x = beq)), nonlinear inequalities ((c(x) \leq 0)), nonlinear equalities ((ceq(x) = 0)), and bound constraints [45].
- PSO: Basic implementations typically handle bound constraints only, though more advanced versions can incorporate various constraint types through penalty functions or specialized techniques [46].
Gradient Utilization and Methodological Approaches
A critical distinction between these optimization methods lies in their use of gradient information:
- lsqnonlin: Implements both large-scale and medium-scale algorithms. The large-scale algorithm uses a subspace trust region method based on the interior-reflective Newton method, while the medium-scale algorithm uses the Levenberg-Marquardt or Gauss-Newton method with line search [44]. Both approaches can utilize gradient information when available.
- fmincon: Offers multiple algorithms including interior-point, sequential quadratic programming (SQP), and active-set methods [45]. For code generation, only the 'sqp' or 'sqp-legacy' algorithms are supported [48].
- PSO: As a metaheuristic, PSO does not use gradient information, making it suitable for problems where gradients are unavailable, unreliable, or computationally expensive to calculate [46].
Table 2: Algorithm Characteristics and Suitable Applications
| Algorithm | Problem Type | Gradient Use | Constraint Handling | Best-Suited Applications |
|---|---|---|---|---|
| lsqnonlin | Nonlinear least-squares | Optional | Bound, linear, nonlinear | Data fitting, curve fitting, parameter estimation with residual minimization [43] |
| fmincon | General constrained nonlinear minimization | Optional | Comprehensive | General optimization with multiple constraint types [45] |
| PSO | General black-box optimization | None | Primarily bound constraints | Non-convex, non-smooth, noisy, or discontinuous problems [46] |
Implementation Protocols and Experimental Considerations
Practical Implementation Guidelines
lsqnonlin Implementation
For effective use of lsqnonlin, the objective function must be formulated to return the vector of residual values (F(x) = [f1(x), f2(x), ..., f_n(x)]) rather than the sum of squares [43] [44]. A typical implementation follows this pattern:
fmincon Implementation
When using fmincon for least-squares problems, the objective function must return the scalar sum of squares:
PSO Implementation
A basic PSO implementation follows this structure [46]:
Workflow for Parameter Estimation Problem Formulation
The following diagram illustrates a systematic workflow for formulating parameter estimation problems and selecting appropriate optimization methods:
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Computational Tools for Optimization-Based Parameter Estimation
| Tool/Resource | Function | Implementation Notes |
|---|---|---|
| MATLAB Optimization Toolbox | Provides lsqnonlin and fmincon solvers |
Requires license; extensive documentation and community support [43] [45] |
| MATLAB Coder | Enables deployment of optimization code to C/C++ | Allows deployment on hardware without MATLAB; requires additional license [48] |
| Automatic Differentiation | Calculates derivatives algorithmically | More accurate and efficient than finite differences; reduces function evaluations [47] |
| NLopt Optimization Library | Alternative open-source optimization library | Includes multiple global and local algorithms; useful for C++ implementations [49] |
| Sensitivity Analysis Tools | Identifiable parameter subset selection | Methods: correlation matrix analysis, SVD with QR, Hessian eigenvector analysis [3] |
| Parameter Subset Selection | Identifies estimable parameters given model and data | Critical for complex models with sparse data; avoids unidentifiable parameters [3] |
Application to Pharmacological and Biological Systems
Parameter estimation in pharmacological and biological systems presents unique challenges, including sparse data, complex nonlinear dynamics, and only partially observable system states [3]. The three methods discussed offer complementary approaches to addressing these challenges:
Case Study: Cardiovascular Model Parameter Estimation
Research on baroreceptor feedback regulation of heart rate during head-up tilt illustrates the practical application of parameter estimation methods [3]. This nonlinear differential equations model contains parameters representing physiological quantities such as afferent neuron gain and neurotransmitter time scales. When applying parameter estimation to such biological systems:
- Identifiability analysis is crucial before estimation, as only a subset of parameters may be estimable given the available data [3]
- Method selection depends on problem structure:
lsqnonlinexcels for residual minimization,fminconfor general constrained problems, and PSO for global exploration of complex parameter spaces - Hybrid approaches can be effective, using PSO for initial global exploration followed by gradient-based methods for refinement [50]
Performance in Cosmological Parameter Estimation
Recent research in cosmology has demonstrated the effectiveness of PSO for estimating cosmological parameters from Type Ia Supernovae and Baryonic Acoustic Oscillations data [50]. These studies show that PSO can deliver competitive results compared to traditional Markov Chain Monte Carlo (MCMC) methods at a fraction of the computational time, and PSO outputs can serve as valuable initializations for MCMC methods to accelerate convergence [50].
Selecting an appropriate optimization method for parameter estimation requires careful consideration of problem structure, data characteristics, and computational constraints. lsqnonlin provides superior efficiency for nonlinear least-squares problems with its specialized handling of residual vectors. fmincon offers greater flexibility for generally constrained optimization problems at the cost of additional function evaluations. PSO serves as a powerful gradient-free alternative for non-convex, noisy, or discontinuous problems where traditional gradient-based methods struggle. A well-formulated parameter estimation strategy should include preliminary identifiability analysis, appropriate algorithm selection based on problem characteristics, and validation of results against experimental data. By understanding the strengths and limitations of each optimization approach, researchers can develop more robust and reliable parameter estimation pipelines for complex scientific and engineering applications.
Formulating a parameter estimation problem is a critical step in translating a biological hypothesis into a quantifiable, testable mathematical model. Within pharmacokinetics (PK), this process is paramount for understanding drug absorption, a process often described by complex, non-linear models. The core thesis of this guide is that a robust parameter estimation problem must be constructed around three pillars: a clearly defined physiological hypothesis, the selection of an identifiable model structure, and the application of appropriate numerical and statistical methods for estimation and validation. This framework moves beyond simple curve-fitting to ensure estimated parameters are reliable, interpretable, and predictive for critical tasks like bioavailability prediction and formulation development [51] [52].
This whitepaper will elucidate this formulation process through a detailed case study estimating the absorption rate constant (k~a~) for a drug following a two-compartment model, where traditional methods requiring intravenous (IV) data are not feasible [51]. We will integrate methodologies from classical PK, physiologically-based pharmacokinetic (PBPK) modeling, and advanced statistical estimation techniques [3] [53].
Theoretical Foundation: Drug Absorption Models
Oral drug absorption is a sequential process involving disintegration, dissolution, permeation, and first-pass metabolism. The choice of mathematical model depends on the dominant rate-limiting steps and available data.
- Compartmental Models: These are the most common in clinical PK analysis. The body is represented as a system of interconnected compartments.
- One-Compartment with First-Order Absorption: A basic model where k~a~ and elimination rate constant (k) can be derived from plasma concentration-time data.
- Two-Compartment with First-Order Absorption: Necessary for drugs displaying distinct distribution and elimination phases. Classical estimation of k~a~ for this model (e.g., Loo-Riegelman method) requires IV data to obtain distribution micro-constants (k~12~, k~21~) and elimination constant (k~10~) [51].
- Physiologically-Based Absorption Models: Models like the Advanced Compartmental Absorption and Transit (ACAT) or convection-dispersion models incorporate physiological parameters (transit times, regional pH, surface area) and drug-specific properties (solubility, permeability). They are powerful for predicting food effects, dose-dependent absorption, and guiding formulation strategy [53].
- Models for Complex Absorption Phenomena: For double-peak profiles (as seen with veralipride), models incorporating parallel first-order inputs, sequential zero-order inputs, or transit compartments are used to capture site-specific or discontinuous absorption [54] [55].
Table 1: Overview of Common Drug Absorption Models and Estimation Challenges
| Model Type | Key Parameters | Typical Estimation Method | Primary Challenge |
|---|---|---|---|
| One-Compartment (First-Order) | k~a~, V, k | Direct calculation from T~max~ and C~max~; nonlinear regression | Oversimplification for many drugs. |
| Two-Compartment (First-Order) | k~a~, V~1~, k~12~, k~21~, k~10~ | Loo-Riegelman method; nonlinear regression | Requires IV data for accurate k~a~ estimation [51]. |
| Transit Compartment | k~a~, Mean Transit Time (MTT), # of compartments (n) | Nonlinear mixed-effects modelling (e.g., NONMEM) | Identifiability of n and MTT; more parameters than lag model [55]. |
| PBPK (ACAT) | Permeability, Solubility, Precipitation time | Parameter sensitivity analysis (PSA) coupled with optimization vs. in vivo data [53]. | Requires extensive in vitro and preclinical input; model identifiability. |
Case Study: Estimating k~a~ Without Intravenous Data
Problem Statement: For a drug exhibiting two-compartment disposition kinetics, how can the absorption rate constant (k~a~) be accurately estimated when ethical or practical constraints preclude conducting an IV study to obtain the necessary disposition parameters (k~12~, k~21~, k~10~)?
Proposed Solution – The "Direct Method": A novel approach defines a new pharmacokinetic parameter: the maximum apparent rate constant of disposition (k~max~). This is the maximum instantaneous rate constant of decline in the plasma concentration after T~max~, occurring at time τ (tau) [51].
Theoretical Development:
In a two-compartment model after extravascular administration, the decline post-T~max~ is not log-linear. The derivative of the log-concentration curve changes over time. The point of steepest decline (most negative slope) is k~max~. The method derives an approximate relationship:
k~a~ / k~max~ ≈ (τ - T~max~) / τ [51].
From this, k~a~ can be solved if k~max~, τ, and T~max~ are known. k~max~ and τ can be determined directly from the oral plasma concentration-time profile by finding the point of maximum negative slope in the post-T~max~ phase, requiring no IV data.
Experimental Protocol for Validation [51]:
- Clinical Study Design: A randomized, cross-over bioequivalence study is conducted with two immediate-release formulations (Test and Reference) of a model drug (e.g., Telmisartan).
- Subjects: Healthy adult volunteers (n ≥ 18), fasted overnight.
- Dosing & Sampling: A single oral dose is administered with water. Serial blood samples are collected pre-dose and at frequent intervals up to 72 hours post-dose to fully characterize the concentration-time curve.
- Bioanalysis: Plasma concentrations are determined using a validated LC-MS/MS method.
- Non-Compartmental Analysis (NCA): Primary PK parameters (AUC~0-t~, AUC~0-∞~, C~max~, T~max~) are calculated using standard methods.
- Application of Direct Method: a. Plot log(plasma concentration) vs. time for each subject. b. Identify T~max~ from the raw concentration data. c. In the post-T~max~ phase, calculate the local slope (derivative) between consecutive points. d. Identify the time point τ corresponding to the most negative slope. This slope is k~max~. e. Calculate k~a~ using the derived formula.
- Validation: The estimated k~a~ values are correlated with absorption-sensitive PK parameters (C~max~/AUC, T~max~) to assess face validity. Accuracy is compared against values estimated by the Loo-Riegelman method (when simulated IV parameters are used) and the statistical moment method.
Table 2: Exemplar Results from Direct Method Application (Telmisartan)
| Formulation | T~max~ (h) | τ (h) | k~max~ (h⁻¹) | Estimated k~a~ (h⁻¹) | C~max~/AUC (h⁻¹) |
|---|---|---|---|---|---|
| FM1 (Reference) | 1.50 (mean) | 4.20 (mean) | 0.25 (mean) | 0.65 (mean) | 0.105 |
| FM2 (Test) | 1.25 (mean) | 3.80 (mean) | 0.28 (mean) | 0.78 (mean) | 0.112 |
The General Parameter Estimation Workflow
Formulating the problem follows a structured workflow applicable across model types, from classical to PBPK.
Diagram 1: Parameter Estimation Formulation Workflow
Step-by-Step Methodology:
- Hypothesis & Model Selection: The hypothesis (e.g., "absorption is first-order and rate-limiting") dictates the model structure. At this stage, structural identifiability should be considered—whether parameters can be uniquely estimated from perfect data [3].
- Experimental Design: The study must provide informative data for the parameters of interest. For absorption, dense sampling during the absorption and early distribution phases is critical [52].
- Problem Formulation (The Optimization Problem): This is the core of the case study. The problem is defined as:
- Parameters (θ): The vector of unknowns to estimate (e.g., θ = [k~a~, V~1~, k~12~, k~21~, k~10~]).
- Cost Function, S(θ): Typically a weighted least squares function:
S(θ) = Σ (Y~i~ - f(t~i~; θ))² / σ², where Y~i~ are observed concentrations, f(t~i~;θ) is the model prediction, and σ² is the variance. - Constraints: Physiological bounds on parameters (e.g., k~a~ > 0).
- Practical Identifiability & Subset Selection: With real, noisy, and sparse data, not all parameters may be practically identifiable. Methods like sensitivity analysis (calculating ∂f/∂θ), correlation matrix analysis, or singular value decomposition (SVD) can identify which parameter subsets can be reliably estimated [3]. For example, in a complex model, k~12~ and k~21~ might be highly correlated and not independently identifiable from oral data alone.
- Numerical Optimization: Algorithms (e.g., Levenberg-Marquardt, SAEM for population data) find the parameter set θ* that minimizes S(θ). For non-convex problems, global optimization techniques may be needed to avoid local minima [4].
- Validation: Use diagnostic plots (observed vs. predicted, residuals) and external validation (e.g., predicting a different dose) [54].
The Scientist's Toolkit: Essential Research Reagents & Solutions
Table 3: Key Reagents and Tools for Absorption Model Parameter Estimation
| Category | Item/Solution | Function in Parameter Estimation |
|---|---|---|
| In Vivo/Clinical | Clinical PK Study Samples (Plasma) | Provides the primary time-series data (Y~i~) for fitting the model. |
| In Vitro Assays | Caco-2 Permeability Assay | Provides an initial estimate for human effective permeability (P~eff~), a key input for PBPK models [53]. |
| Equilibrium Solubility (Multiple pH) | Measures drug solubility, critical for predicting dissolution-limited absorption, especially for BCS Class II drugs [53]. | |
| Software & Algorithms | Nonlinear Mixed-Effects Modeling Software (NONMEM, Monolix) | Industry standard for population PK analysis, capable of fitting complex models (transit, double absorption) to sparse data [54] [55]. |
| PBPK/Simulation Platforms (GastroPlus, SIMCYP, PK-SIM) | Implement mechanistic absorption models (ACAT, dispersion). Used for Parameter Sensitivity Analysis (PSA) to guide formulation development and assess identifiability [53] [56]. | |
| Global Optimization Toolboxes (e.g., αBB) | Solve non-convex estimation problems to find global parameter minima, reducing dependency on initial estimates [4]. | |
| Parameter Identifiability Analysis Tools (e.g., COMBOS, custom SVD scripts) | Perform structural and practical identifiability analysis to define estimable parameter subsets before fitting [3]. | |
| Mathematical Constructs | Sensitivity Matrix (∂f/∂θ) | Quantifies how model outputs change with parameters; basis for identifiability analysis and optimal design [3]. |
| Statistical Moment Calculations (AUC, AUMC, MRT) | Provides non-compartmental estimates used in methods like the statistical moment method for k~a~ [51]. |
Visualizing Model Structures and Relationships
Diagram 2: Two-Compartment PK Model with Oral Input
Diagram 3: Direct Method Algorithm for k~a~ Estimation
This case study demonstrates that formulating a parameter estimation problem is a deliberate exercise in balancing biological reality with mathematical and practical constraints. The "Direct Method" for estimating k~a~ exemplifies an innovative solution crafted around a specific data limitation. The broader thesis underscores that successful formulation requires: 1) a model reflecting key physiology, 2) an experimental design that informs it, 3) a rigorous assessment of which parameters can actually be gleaned from the data (identifiability), and 4) robust numerical and statistical methods for estimation. By adhering to this framework—supported by tools ranging from sensitivity analysis to global optimization—researchers can develop predictive absorption models that reliably inform drug development decisions, from formulation screening to bioequivalence assessment [51] [53] [56].
Overcoming Practical Hurdles: Strategies for Complex Models and Noisy Data
Parameter estimation is a fundamental inverse problem in computational biology, critical for transforming generic mathematical models into patient- or scenario-specific predictive tools [3]. This process involves determining the values of model parameters that enable the model outputs to best match experimental observations. For complex biological systems, such as those described by nonlinear differential equations or whole-cell models, parameter estimation presents significant computational challenges [3] [57]. Models striving for physiological comprehensiveness often incorporate numerous parameters, many of which are poorly characterized or unknown, creating a high-dimensional optimization landscape [57].
The core challenge lies in the computational expense of evaluating complex models. As noted in the DREAM8 Whole-Cell Parameter Estimation Challenge, a single simulation of the Mycoplasma genitalium whole-cell model can require approximately one core day to complete [57]. This computational burden renders traditional optimization approaches, which may require thousands of model evaluations, practically infeasible. Furthermore, issues of structural and practical identifiability complicate parameter estimation, as not all parameters can be reliably estimated given limited and noisy experimental data [3]. These challenges have motivated the development of sophisticated approaches, including hybrid methods and model reduction techniques, to make parameter estimation tractable for complex biological systems.
Foundational Concepts and Definitions
The Parameter Estimation Problem
The parameter estimation problem can be formally defined for models described by nonlinear systems of equations. Given a model state vector x ∈ Rⁿ and parameter vector θ ∈ R^q, the system dynamics are described by:
dx/dt = f(t, x; θ) (1)
with an output vector y ∈ R^m corresponding to available measurements:
y = g(t, x; θ) (2)
The inverse problem involves finding the parameter values θ that minimize the difference between model outputs and experimental data Y sampled at specific time points [3].
Key Computational Challenges
- High-Dimensional Parameter Spaces: Whole-cell models may contain thousands of parameters, creating a complex optimization landscape [57].
- Nonconvex Objective Functions: The nonlinear nature of biological models often results in objective functions with multiple local minima [4].
- Structural and Practical Unidentifiability: It may be impossible to estimate all parameters given the model structure and available data [3].
- Computational Expense: Complex hybrid models can be prohibitively slow to evaluate, limiting the applicability of traditional optimization techniques [57].
Table 1: Classification of Parameter Estimation Challenges
| Challenge Type | Description | Impact on Estimation |
|---|---|---|
| Structural Identifiability | Inability to determine parameters due to model structure | Fundamental limitation requiring model reformulation |
| Practical Identifiability | Insufficient data quality or quantity for reliable estimation | May be addressed through improved experimental design |
| Computational Complexity | High computational cost of model simulations | Limits application of traditional optimization methods |
| Numerical Stability | Sensitivity to initial conditions and parameter scaling | Can prevent convergence to optimal solutions |
Model Reduction Techniques
Principles of Model Reduction
Model reduction techniques address computational challenges by replacing original complex models with simpler approximations that retain essential dynamical features. These techniques minimize the computational cost of optimization by creating cheaper, approximate functions that can be evaluated more rapidly than the original model [57]. In the context of parameter estimation, reduced models serve as surrogates during the optimization process, allowing for more extensive parameter space exploration.
Karr et al. employed a reduction approach for whole-cell model parameterization by constructing "a reduced physical model that approximates the temporal and population average of the full model" [57]. This reduced model maintained the same parameters as the full model but was computationally cheaper to evaluate, enabling parameter optimization that would have been infeasible with the full model alone.
Implementation Methodology
The implementation of model reduction for parameter estimation follows a structured workflow:
- Model Analysis: Identify critical dynamics and parameters that dominate system behavior.
- Reduction Technique Selection: Choose appropriate reduction methods based on model characteristics.
- Surrogate Model Construction: Create simplified models that approximate key behaviors.
- Parameter Optimization: Estimate parameters using the reduced model.
- Validation: Verify parameter values with the original full-scale model.
Table 2: Model Reduction Techniques for Parameter Estimation
| Technique | Mechanism | Applicable Model Types | Computational Savings |
|---|---|---|---|
| Timescale Separation | Explores disparities in reaction rates | Biochemical networks, metabolic systems | High (reduces stiffness) |
| Sensitivity Analysis | Identifies most influential parameters | Large-scale biological networks | Medium (reduces parameter space) |
| Principal Component Analysis | Projects dynamics onto dominant modes | High-dimensional systems | Variable (depends on dimension reduction) |
| Balanced Truncation | Eliminates weakly controllable/observable states | Linear and weakly nonlinear systems | High for appropriate systems |
| Proper Orthogonal Decomposition | Uses empirical basis functions from simulation data | Nonlinear parameterized systems | Medium to high |
Hybrid Methods for Parameter Estimation
Framework for Hybrid Optimization
Hybrid methods combine multiple optimization strategies to leverage their respective strengths while mitigating individual weaknesses. These approaches are particularly valuable for addressing the nonconvex nature of parameter estimation problems in nonlinear biological models [4]. The global optimization approach based on a branch-and-bound framework presented by Adjiman et al. represents one such hybrid strategy for solving the error-in-variables formulation in nonlinear algebraic models [4].
Hybrid methods typically integrate:
- Global Exploration Strategies: Techniques such as genetic algorithms, particle swarm optimization, or branch-and-bound methods that broadly explore parameter space.
- Local Refinement Algorithms: Gradient-based methods like Levenberg-Marquardt or trust-region approaches that converge efficiently to local minima.
- Surrogate-Assisted Optimization: Machine learning models including artificial neural networks, splines, and support vector machines that approximate the objective function [57].
Distributed Optimization Framework
Distributed optimization represents a powerful hybrid approach that uses "multiple agents, each simultaneously employing the same algorithm on different regions, to quickly identify optima" [57]. In this framework, agents typically cooperate by exchanging information, allowing them to learn from each other's experiences and collectively navigate the parameter space more efficiently than single-agent approaches.
Automatic Differentiation in Hybrid Frameworks
Automatic differentiation provides an efficient technique for computing derivatives of computational models by decomposing them into elementary functions and applying the chain rule [57]. This approach enables the use of gradient-based optimization methods for models where finite difference calculations would be prohibitively expensive. When integrated into hybrid frameworks, automatic differentiation enhances local refinement capabilities while maintaining computational feasibility.
Table 3: Hybrid Method Components and Their Roles in Parameter Estimation
| Method Component | Primary Function | Advantages | Implementation Considerations |
|---|---|---|---|
| Global Optimization | Broad exploration of parameter space | Avoids convergence to poor local minima | Computationally intensive; requires careful termination criteria |
| Local Refinement | Efficient convergence to local optima | Fast local convergence; utilizes gradient information | Sensitive to initial conditions; may miss global optimum |
| Surrogate Modeling | Approximates expensive model evaluations | Dramatically reduces computational cost | Introduces approximation error; requires validation |
| Distributed Computing | Parallel evaluation of parameter sets | Reduces wall-clock time; explores multiple regions | Requires communication overhead; implementation complexity |
Experimental Protocols and Case Studies
DREAM8 Parameter Estimation Challenge Protocol
The DREAM8 Whole-Cell Parameter Estimation Challenge provides a robust case study for evaluating hybrid methods and model reduction techniques [57]. The experimental protocol was designed to mimic real-world parameter estimation scenarios:
Challenge Design: Participants were tasked with identifying 15 modified parameters in a whole-cell model of M. genitalium that controlled RNA polymerase promoter binding probabilities, RNA half-lives, and metabolic reaction turnover numbers.
Data Provision: Teams received the model structure, wild-type parameter values, and mutant strain in silico "experimental" data.
Perturbation Allowances: Participants could obtain limited perturbation data to mimic real-world experimental resource constraints.
Evaluation Framework: The competition was divided into four subchallenges, requiring teams to share methodologies to foster collaboration.
Ten teams participated in the challenge, employing varied parameter estimation approaches. The most successful strategies combined multiple techniques to address different aspects of the problem, demonstrating the practical value of hybrid approaches for complex biological models.
Protocol for Parameter Subset Selection
For models with large parameter sets, identifying estimable parameter subsets is crucial. The structured correlation method, singular value decomposition with QR factorization, and subspace identification approaches have been systematically compared for this purpose [3]. The experimental protocol involves:
Sensitivity Matrix Calculation: Compute the sensitivity of model outputs to parameter variations.
Correlation Analysis: Analyze parameter correlations to identify potentially unidentifiable parameters.
Subset Selection: Apply structured correlation analysis, SVD with QR factorization, or subspace identification to select estimable parameter subsets.
Validation: Verify subset selection through synthetic data studies before application to experimental data.
In cardiovascular model applications, the "structured analysis of the correlation matrix" provided the best parameter subsets, though with higher computational requirements than alternative methods [3].
Research Reagent Solutions
Table 4: Essential Computational Tools for Parameter Estimation Research
| Tool/Category | Function | Application in Parameter Estimation |
|---|---|---|
| Global Optimization Algorithms (αBB) | Provides rigorous global optimization for twice-differentiable problems | Identifies global optima in nonconvex parameter estimation problems [4] |
| Sensitivity Analysis Tools | Quantifies parameter influence on model outputs | Identifies sensitive parameters for focused estimation; determines identifiable parameter subsets [3] |
| Automatic Differentiation Libraries | Computes exact derivatives of computational models | Enables efficient gradient-based optimization for complex models [57] |
| Surrogate Modeling Techniques | Creates approximate, computationally efficient model versions | Accelerates parameter estimation through response surface modeling [57] |
| Distributed Computing Frameworks | Enables parallel evaluation of parameter sets | Reduces solution time through concurrent parameter space exploration [57] |
Implementation Guidelines
Formulating the Parameter Estimation Problem
Effective formulation of parameter estimation problems within a research context requires careful consideration of both mathematical and practical constraints:
Identifiability Analysis: Before estimation, assess whether parameters can be uniquely identified from available data. Techniques include structural identifiability analysis using differential algebra and practical identifiability using profile likelihood [3].
Objective Function Design: Formulate objective functions that balance fitting accuracy with regularization terms to address ill-posedness.
Experimental Design: Identify measurement types and sampling protocols that maximize information content for parameter estimation, especially when resources are limited [57].
Computational Budget Allocation: Determine appropriate trade-offs between estimation accuracy and computational resources, particularly for expensive models.
Integration of Hybrid Approaches
Successful implementation of hybrid methods requires strategic integration of complementary techniques:
Problem Decomposition: Divide the parameter estimation problem based on timescales, parameter sensitivities, or model components.
Method Selection: Choose appropriate techniques for different aspects of the problem, matching method strengths to specific challenges.
Information Exchange Protocols: Establish mechanisms for different components of hybrid methods to share information and guide the overall search process.
Termination Criteria: Define multi-faceted convergence criteria that consider both optimization progress and computational constraints.
The integration of these approaches within a coherent framework enables researchers to address parameter estimation challenges that would be intractable with individual methods alone, advancing the broader goal of creating predictive models for complex biological systems.
Parameter estimation is a fundamental process in scientific modeling whereby unknown model parameters are estimated by matching model outputs to observed data (calibration targets) [58]. A critical property of a well-formulated estimation problem is identifiability—the requirement that a unique set of parameter values yields the best fit to the chosen calibration targets [58]. Non-identifiability arises when multiple, distinct parameter sets produce an equally good fit to the available data [58]. This presents a significant challenge for research, as different yet equally plausible parameter values can lead to different scientific conclusions and practical recommendations.
In the context of a broader research thesis, recognizing and resolving non-identifiability is not merely a technical step but a core aspect of ensuring that model-based inferences are reliable and actionable. This is particularly crucial in fields like drug development and systems biology, where models inform critical decisions [59] [58]. Non-identifiability can be broadly categorized as structural or practical. Structural non-identifiability is an intrinsic model property where different parameter combinations yield identical model outputs, creating a true one-to-many mapping from parameters to the data distribution [60]. Practical non-identifiability, on the other hand, arises from limitations in the available data (e.g., noise, sparsity), making it impossible to distinguish between good-fitting parameter values even if the model structure is theoretically identifiable [59] [60].
A Foundational Example: The Signaling Cascade Model
To illustrate the concepts and their implications, consider a four-step biochemical signaling cascade with negative feedback, a motif common in pathways like MAPK (RAS → RAF → MEK → ERK) [59]. This model is typically described by a system of ordinary differential equations with multiple kinetic parameters and feedback strengths.
Diagram 1: A four-step signaling cascade with negative feedback.
A study investigating this cascade demonstrated that training the model using only data for the final variable (K4) resulted in a model that could accurately predict K4's trajectory under new stimulation protocols, even though all 9 model parameters remained uncertain over about two orders of magnitude [59]. This is a classic sign of a sloppy or non-identifiable model. However, this model failed to predict the trajectories of the upstream variables (K1, K2, K3), for which the prediction bands were very broad [59]. Only by sequentially including data for more variables (e.g., K2, then all four) could the model become "well-trained" and capable of predicting all states, effectively reducing the dimensionality of the plausible parameter space [59].
Implications for Decision Making
The practical consequences of non-identifiability are profound. In a separate, simpler example of a three-state Markov model for cancer relative survival, non-identifiability led to two different, best-fitting parameter sets [58]. When used to evaluate a hypothetical treatment, these different parameter sets produced substantially different estimates of life expectancy gain (0.67 years vs. 0.31 years) [58]. This discrepancy could directly influence the perceived cost-effectiveness of a treatment and, ultimately, the optimal decision made by healthcare providers or policymakers [58]. Therefore, checking for non-identifiability is not an academic exercise but a necessary step for robust decision-making.
Detecting and Diagnosing Non-Identifiability
Several robust methodologies exist to diagnose non-identifiability in a model calibration problem.
Profile Likelihood Analysis
This method involves systematically varying one parameter while re-optimizing all others to find the best possible fit to the data. For an identifiable parameter, the profile likelihood will show a distinct, peaked minimum. In contrast, a flat or bimodal profile likelihood indicates that the parameter is not uniquely determined by the data, revealing non-identifiability [58].
Correlation Analysis
High correlations between parameter estimates (e.g., absolute correlation coefficients > 0.98) can indicate that changes in one parameter can be compensated for by changes in another, leading to the same model output [61]. This parameter compensation is a hallmark of non-identifiability, though it's important to note this is a property of the model and data fit, not necessarily the underlying biological mechanism [61].
Collinearity Analysis
This approach is more formal and uses the eigenvalues of the model's Hessian matrix (or an approximation like the Fisher Information Matrix) [3]. A collinearity index is computed; a high index (e.g., above about 3.5) for a parameter subset suggests that the parameters are linearly dependent in their influence on the model output, confirming non-identifiability [58].
Dimensionality Analysis of the Plausible Parameter Space
By performing principal component analysis (PCA) on the logarithms of plausible parameter sets (e.g., from a Markov Chain Monte Carlo sample), one can quantify the effective dimensionality of the parameter space that is constrained by data [59]. A large reduction in the multiplicative deviation (δ) along certain principal components indicates stiff directions that are well-constrained, while many sloppy directions with δ near 1 indicate non-identifiability [59].
Table 1: Methods for Diagnosing Non-Identifiability
| Method | Underlying Principle | Interpretation of Non-Identifiability | Key Advantage |
|---|---|---|---|
| Profile Likelihood [58] | Optimizes all other parameters for each value of a focal parameter. | A flat or biminal likelihood profile. | Intuitive visual output. |
| Correlation Analysis [61] | Computes pairwise correlations from the variance-covariance matrix. | Correlation coefficients near ±1 (e.g., >0.98). | Easy and fast to compute. |
| Collinearity Index [58] | Analyzes linear dependence via the Hessian matrix eigenvalues. | A high index value (demonstrated >3.5). | Formal test for linear dependencies. |
| PCA on Parameter Space [59] | Analyzes the geometry of high-likelihood parameter regions. | Many "sloppy directions" with little change in model output. | Reveals the effective number of identifiable parameter combinations. |
Diagram 2: A workflow for diagnosing non-identifiability during model calibration.
Strategies for Resolving Non-Identifiability
Once non-identifiability is detected, several strategies can be employed to resolve or manage the problem.
Incorporate Additional Calibration Targets
The most direct way to achieve identifiability is to include more informative data. In the cancer survival model example, adding the ratio between the two non-death states over time as an additional calibration target, alongside relative survival, resolved the non-identifiability, resulting in a unimodal likelihood profile and a low collinearity index [58]. The choice of target is critical; measuring a single variable may only constrain a subset of parameters, while successively measuring more variables further reduces the dimensionality of the unidentified parameter space [59].
Optimal Experiment Design (OED) and Bayesian Active Learning
Rather than collecting data arbitrarily, these methods aim to design experiments that will be maximally informative for estimating the uncertain parameters [60]. This is a resource-efficient approach to overcoming practical non-identifiability by targeting data collection to specifically reduce parameter uncertainty.
Model Reduction and Reparametrization
If a model is structurally non-identifiable, it can sometimes be reparameterized into an identifiable form by combining non-identifiable parameters into composite parameters [59]. While this can solve the identifiability problem, a potential drawback is that the resulting composite parameters may lack a straightforward biological interpretation [59].
Parameter Subset Selection and Regularization
When it is not possible to estimate all parameters, one can focus on estimating only an identifiable subset. Methods like analyzing the correlation matrix, singular value decomposition followed by QR factorization, and identifying the subspace closest to the eigenvectors of the model Hessian can help select such a subset [3]. Alternatively, regularization techniques (e.g., L2 regularization or ridge regression) can be used to introduce weak prior information, penalizing unrealistic parameter values and guiding the estimation toward a unique solution [60].
Sequential Training and Analysis of Predictive Power
Instead of insisting on full identifiability, one can adopt a pragmatic, iterative approach. The model is trained on available data, and its predictive power is assessed for specific, clinically or biologically relevant scenarios, even if some parameters remain unknown [59]. For example, a model trained only on the last variable of a cascade might still accurately predict that variable's response to different stimuli, which could be sufficient for a particular application [59]. Subsequent experiments can then be designed based on the model's current predictive limitations.
Table 2: Strategies for Resolving Non-Identifiability
| Strategy | Description | Best Suited For | Considerations |
|---|---|---|---|
| Additional Data [58] | Incorporating more or new types of calibration targets. | Practical non-identifiability. | Can be costly or time-consuming; OED optimizes this. |
| Model Reduction [59] | Combining parameters or simplifying the model structure. | Structural non-identifiability. | May result in loss of mechanistic interpretation. |
| Subset Selection [3] | Identifying and estimating only a subset of identifiable parameters. | Complex models with many parameters. | Leaves some parameters unknown; requires specialized methods. |
| Regularization (L2) [60] | Adding a penalty to the likelihood to constrain parameter values. | Ill-posed problems and practical non-identifiability. | Introduces prior bias; solution depends on penalty strength. |
| Sequential Training [59] | Iteratively training the model and assessing its predictive power. | Resource-limited settings where full data is unavailable. | Accepts uncertainty in parameters but validates useful predictions. |
Diagram 3: An iterative, sequential approach to model training and validation when faced with non-identifiability.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Key Research Reagent Solutions for Parameter Estimation Studies
| Reagent / Material | Function in Experimental Protocol |
|---|---|
| Optogenetic Receptors [59] | Allows application of complex, temporally precise stimulation protocols to perturb the biological system and generate informative data for constraining dynamic models. |
| Markov Chain Monte Carlo (MCMC) Algorithms [59] | A computational method for sampling the posterior distribution of model parameters, enabling exploration of the plausible parameter space and assessment of identifiability. |
| Sensitivity & Correlation Analysis Software [3] | Tools to compute parameter sensitivities, correlation matrices, and profile likelihoods, which are essential for diagnosing practical non-identifiability. |
| Global Optimization Algorithms (e.g., αBB) [4] | Computational methods designed to find the global minimum of nonconvex objective functions, helping to avoid local minima during parameter estimation. |
| Case-Specific Biological Assays | Assays to measure intermediate model variables (e.g., K1, K2, K3 in the cascade) are crucial for successive model constraint and resolving non-identifiability [59]. |
Non-identifiability is a fundamental challenge in parameter estimation that, if unaddressed, can severely undermine the validity of model-based research. It is imperative to formally check for its presence using methods like profile likelihood, correlation, and collinearity analysis. The strategies for dealing with non-identifiability form a continuum, from collecting additional data and redesigning experiments to reducing model complexity and adopting a pragmatic focus on the predictive power for specific tasks. For researchers formulating a parameter estimation problem, a systematic approach that includes identifiability assessment as a core component is not optional but essential for producing trustworthy, reproducible, and actionable scientific results.
Systematic model-based design of experiment (MBDoE) is essential to maximise the information obtained from experimental campaigns, particularly for systems described by stochastic or non-linear models where information quantity is characterized by intrinsic uncertainty [62]. This technique becomes critically important when designing experiments to estimate parameters in complex biological, pharmacological, and chemical systems where traditional one-factor-at-a-time approaches prove inadequate. The fundamental challenge in parameter estimation lies in maximizing the information content of experimental data while minimizing resource requirements and experimental burden [63].
The process of optimizing experimental design for parameter estimation represents an inverse problem where we seek to determine the model parameters that best describe observed data [63]. In non-linear systems, which commonly represent biological and chemical processes through ordinary differential equations, this task is particularly challenging. The optimal design depends on the "true model parameters" that govern the system evolution—precisely what we aim to discover through experimentation [63]. This circular dependency necessitates sophisticated approaches that can sequentially update parameter knowledge through iterative experimental campaigns.
Within the context of a broader thesis on formulating parameter estimation problems, this guide establishes the mathematical foundations, methodological frameworks, and practical implementation strategies for designing experiments that yield maximally informative data for parameter estimation. By focusing on the selection of operating profiles (experimental conditions) and data points (sampling strategies), we provide researchers with a systematic approach to overcoming the identifiability challenges that plague complex model development.
Theoretical Foundations
The Parameter Estimation Problem
Parameter estimation begins with a mathematical model representing system dynamics. Biological quantities such as molecular concentrations are represented by states x(t) and typically follow ordinary differential equations:
ẋ(t) = f(x, p, u)
where the function f defines the system dynamics, p represents the unknown model parameters to be estimated, and u represents experimental perturbations or controls [63]. The observables y(t) = g(x(t), s_obs) + ϵ represent the measurable outputs, where ϵ accounts for observation noise [63].
The parameter estimation problem involves finding the parameter values p that minimize the difference between model predictions and experimental data. For non-linear models, this typically requires numerical optimization methods to maximize the likelihood function or minimize the sum of squared residuals [63]. The quality of these parameter estimates depends critically on the experimental conditions under which data are collected and the sampling points selected for measurement.
Information Matrices in Experimental Design
The Fisher Information Matrix (FIM) serves as a fundamental tool for quantifying the information content of an experimental design. For a parameter vector θ, the FIM is defined as:
FIM = E[(∂ log L/∂θ) · (∂ log L/∂θ)^T]
where L is the likelihood function of the parameters given the data [64]. The inverse of the FIM provides an approximation of the parameter covariance matrix, establishing a direct link between experimental design and parameter uncertainty [63].
In practical applications, optimal experimental design involves optimizing some function of the FIM. Common optimality criteria include:
- D-optimality: Maximizes the determinant of the FIM, minimizing the volume of the confidence ellipsoid
- A-optimality: Minimizes the trace of the inverse FIM, reducing average parameter variance
- E-optimality: Maximizes the minimum eigenvalue of the FIM, improving the worst-case parameter direction
For non-linear systems, the FIM depends on the parameter values themselves, creating the circular dependency that sequential experimental design strategies aim to resolve [63].
Table 1: Optimality Criteria for Experimental Design
| Criterion | Objective | Application Context |
|---|---|---|
| D-optimality | Maximize determinant of FIM | General purpose; minimizes overall confidence region volume |
| A-optimality | Minimize trace of inverse FIM | Focus on average parameter variance |
| E-optimality | Maximize minimum eigenvalue of FIM | Improve worst-case parameter direction |
| Modified E-optimality | Minimize condition number of FIM | Improve parameter identifiability |
Methodological Approaches
Stochastic Model-Based Design of Experiments (SMBDoE)
The SMBDoE approach represents an advanced methodology that simultaneously identifies optimal operating conditions and allocation of sampling points in time [62]. This method is particularly valuable for systems with significant intrinsic stochasticity, where uncertainty characterization fundamentally impacts experimental design decisions.
SMBDoE employs two distinct sampling strategies that select sampling intervals based on different characteristics of the Fisher information:
- Average Information Strategy: Focuses on the expected value of Fisher information across parameter distributions
- Uncertainty-Aware Strategy: Explicitly incorporates the uncertainty in Fisher information resulting from system stochasticity
This approach acknowledges that in stochastic systems, the information quantity itself is uncertain, and this uncertainty should influence experimental design decisions [62]. By accounting for this uncertainty, SMBDoE generates more robust experimental designs that perform well across the range of possible system behaviors.
Two-Dimensional Profile Likelihood Approach
For non-linear systems where the FIM may inadequately represent parameter uncertainty, the two-dimensional profile likelihood approach offers a powerful alternative [63]. This method quantifies the expected uncertainty of a targeted parameter after a potential measurement, providing a design criterion that meaningfully represents expected parameter uncertainty reduction.
The methodology works as follows:
- For a specified experimental condition, compute the two-dimensional profile likelihood that accounts for different possible measurement outcomes
- Quantify the range of reasonable measurement outcomes using validation profiles
- Determine the parameter uncertainty after each specific measurement outcome via respective profile likelihoods
- Define a design criterion representing the expected average width of the confidence interval after measuring data for the experimental condition
This approach effectively reverses the standard profile likelihood logic: instead of assessing how different parameters affect model predictions, it evaluates how different measurement outcomes will impact parameter estimates [63]. The resulting two-dimensional likelihood profiles serve as both quantitative design tools and intuitive visualizations of experiment impact.
Accounting for Observation Noise
The presence and structure of observation noise significantly impacts optimal experimental design. Research demonstrates that correlations in observation noise can dramatically alter the optimal time points for system observation [64]. Proper consideration of observation noise must therefore be integral to the experimental design process.
Methods that combine local sensitivity measures (from FIM) with global sensitivity measures (such as Sobol' indices) provide a comprehensive framework for designing experiments under observation noise [64]. The optimization of observation times must explicitly incorporate noise characteristics to achieve minimal parameter uncertainty.
Figure 1: Incorporating observation noise characteristics into experimental design optimization
Implementation Protocols
Sequential Experimental Design Workflow
Implementing optimal experimental design requires a structured workflow that integrates modeling, design optimization, experimentation, and analysis. The following protocol outlines a sequential approach that progressively reduces parameter uncertainty:
Figure 2: Sequential workflow for iterative experimental design and parameter estimation
Protocol for Optimal Sampling Time Selection
Selecting informative sampling time points represents a critical aspect of experimental design. The following step-by-step methodology enables researchers to determine optimal observation times:
- Define Model and Parameters: Establish the mathematical model structure and identify parameters for estimation
- Specify Experimental Constraints: Define practical limitations (total experiment duration, minimum time between samples, maximum number of samples)
- Compute Sensitivity Coefficients: Calculate the sensitivity of model outputs to parameter variations: Si(t) = ∂y(t)/∂θi
- Formulate Design Criterion: Select appropriate optimality criterion (D-optimal, A-optimal, etc.) based on research objectives
- Optimize Sampling Times: Solve the optimization problem to determine time points that maximize the design criterion
- Validate Design: Perform simulation studies to verify expected performance
This protocol emphasizes that optimal sampling strategies depend not only on system dynamics but also on the specific parameters of interest and the noise characteristics of the measurement process [62] [64].
Protocol for Operating Condition Selection
Choosing optimal operating conditions (e.g., temperature, pH, initial concentrations, stimulus levels) follows a complementary procedure:
- Identify Manipulable Variables: Determine which input variables u can be practically controlled
- Define Operating Ranges: Establish feasible ranges for each manipulable variable based on practical constraints
- Parameterize Input Profiles: Represent input trajectories using appropriate parameterization (step functions, polynomials, etc.)
- Compute Profile Sensitivities: Evaluate how parameter uncertainties depend on input profiles
- Optimize Operating Conditions: Determine input profiles that maximize information content for parameter estimation
In both sampling time and operating condition selection, the sequential nature of optimal experimental design means that initial experiments may be designed based on preliminary parameter estimates, with subsequent experiments designed using updated estimates [63].
Table 2: Experimental Design Optimization Tools and Their Applications
| Methodology | Key Features | Implementation Considerations |
|---|---|---|
| Fisher Information Matrix (FIM) | Linear approximation, computationally efficient | May perform poorly with strong non-linearity or limited data |
| Profile Likelihood-Based | Handles non-linearity well, more computationally intensive | Better for small to medium parameter sets |
| Stochastic MBDoE | Explicitly accounts for system stochasticity | Requires characterization of uncertainty sources |
| Two-Dimensional Profile Likelihood | Visualizes experiment impact, handles non-linearity | Computationally demanding for large systems |
Applications in Drug Development
Model-Informed Drug Development (MIDD)
The pharmaceutical industry has embraced model-based experimental design through Model-Informed Drug Development (MIDD), which provides a quantitative framework for advancing drug development and supporting regulatory decision-making [10]. MIDD plays a pivotal role throughout the drug development lifecycle:
- Early Discovery: Target identification and lead compound optimization through QSAR modeling
- Preclinical Development: First-in-human dose prediction using PBPK and allometric scaling
- Clinical Development: Optimization of clinical trial designs through adaptive approaches and population PK/PD modeling
- Post-Market: Support for label updates and additional indication approvals
The "fit-for-purpose" principle guides MIDD implementation, ensuring that modeling approaches align with specific questions of interest and context of use at each development stage [10]. This strategic application of modeling and simulation maximizes information gain while minimizing unnecessary experimentation.
Clinical Trial Simulations
Clinical Trial Simulation (CTS) represents a powerful application of optimal experimental design in drug development [65]. CTS uses computer programs to mimic clinical trial conduct based on pre-specified models that reflect the actual situation being simulated. The primary objective is to describe, extrapolate, or predict clinical trial outcomes, enabling researchers to:
- Investigate assumptions in clinical trial designs
- Influence trial design to maximize pertinent information
- Study disease pathogenesis and progression
- Evaluate new treatment strategies
- Select optimal doses and dosing regimens
- Maximize chances of clinical trial success
For AIDS research, for example, CTS incorporates mathematical models for pharmacokinetics/pharmacodynamics of antiviral agents, adherence, drug resistance, and antiviral responses [65]. This integrated approach enables more informative clinical trial designs that efficiently address key research questions.
Practical Considerations and Research Reagents
Research Reagent Solutions for Experimental Implementation
Table 3: Essential Research Reagents and Computational Tools for Implementation
| Resource Category | Specific Examples | Function in Experimental Design |
|---|---|---|
| Modeling Software | MATLAB with Data2Dynamics toolbox [63], R with dMod package | Parameter estimation, profile likelihood computation, sensitivity analysis |
| Optimization Tools | Global optimization algorithms, Markov Chain Monte Carlo samplers | Design criterion optimization, uncertainty quantification |
| Simulation Platforms | Clinical Trial Simulation software [65], PBPK modeling tools | Virtual experiment evaluation, design validation |
| Laboratory Equipment | Automated sampling systems, precise environmental control | Implementation of optimized sampling times and operating conditions |
Protocol Documentation Standards
Comprehensive protocol documentation ensures experimental consistency and reproducibility. Effective research protocols should include [66]:
- Metadata: Protocol title, keywords, authors, and description providing context
- Step-by-Step Instructions: Granular, clearly explained procedures
- Checklists: Traceable verification of critical steps
- Tables and Calculations: Structured presentation of experimental conditions
- Attachment: Relevant files, plate maps, or instrument instructions
Well-structured protocols balance completeness with conciseness, breaking complex procedures into simpler steps to reduce errors and enhance reproducibility [67]. This documentation practice is essential for maintaining experimental rigor throughout iterative design processes.
Optimizing experimental design through systematic selection of operating profiles and data points represents a powerful methodology for enhancing parameter estimation in complex systems. By integrating approaches ranging from Fisher information-based methods to advanced profile likelihood techniques, researchers can dramatically improve the information content of experimental data. The sequential application of these methods—designing each experiment based on knowledge gained from previous iterations—enables efficient parameter estimation even for highly non-linear systems with practical constraints.
The implementation of these strategies within structured frameworks like Model-Informed Drug Development demonstrates their practical utility across scientific domains. As computational power increases and methodological innovations continue emerging, optimal experimental design will play an increasingly vital role in maximizing knowledge gain while minimizing experimental burden across scientific discovery and product development.
Leveraging Machine Learning and Data Assimilation to Constrain Model Parameters
Parameter estimation is a fundamental process in computational science, crucial for building models that accurately represent real-world systems across diverse fields. The core challenge lies in tuning a model's internal parameters—values not directly observable—so that its output faithfully matches observed data. This process of "constraining" or "calibrating" a model is essential for ensuring its predictive power and reliability. In complex systems, from drug interactions to climate models, parameters often cannot be measured directly and must be inferred from their effects on observable outcomes.
Traditional statistical and optimization methods, while effective for simpler systems, often struggle with the high-dimensional, noisy, and non-linear problems common in modern science. The emergence of machine learning (ML) and sophisticated data assimilation (DA) techniques has revolutionized this field. ML excels at identifying intricate, non-linear relationships from large, noisy datasets, while DA provides a rigorous mathematical framework for dynamically integrating observational data with model forecasts to improve state and parameter estimates. This guide explores the integrated ML-DA methodology, providing researchers with the technical foundation to formulate and solve advanced parameter estimation problems, thereby enhancing model accuracy and predictive capability in their respective domains.
Theoretical Foundations
The Core Parameter Estimation Problem
At its heart, parameter estimation is an inverse problem. Given a model and a set of observations, the goal is to find the parameter values that minimize the discrepancy between the model's prediction and the observed data.
A dynamical system is often described by an Ordinary Differential Equation (ODE): $$\frac{d\textbf{x}(t)}{dt} = f(\textbf{x}(t), t, \theta)$$ where $\textbf{x}(t)$ is the system state at time $t$, $f$ is the function governing the system dynamics, and $\theta$ represents the parameters to be estimated [68].
The estimation process typically involves defining a loss function (or cost function) that quantifies this discrepancy. Traditional methods like Nonlinear Least Squares (NLS) aim to find parameters $\theta$ that minimize the sum of squared residuals: $\min\theta \sum{i=1}^N ||yi - Mi(\theta)||^2$, where $yi$ are observations and $Mi(\theta)$ are model predictions [68]. However, such methods can be highly sensitive to noise and model imperfections.
Machine Learning for Parameter Estimation
Machine learning offers a powerful, data-driven alternative to traditional methods. ML models, particularly neural networks, are inherently designed to learn complex mappings from data without requiring pre-specified formulas relating inputs to outputs [69]. This makes them exceptionally well-suited for parameter estimation where the underlying relationships are complex or poorly understood.
A key advancement is the use of robust loss functions like the Huber loss, which combines the advantages of Mean Squared Error (MSE) and Mean Absolute Error (MAE). This makes the estimation process more resilient to outliers and noise, which are common in experimental data [68]. Studies have demonstrated that neural networks employing Huber loss can maintain sub-1.2% relative errors in key parameters even for chaotic systems like the Lorenz model, significantly outperforming NLS, which can diverge with errors exceeding 12% under identical noise conditions [68].
Data Assimilation for Parameter Constraints
Data Assimilation provides a Bayesian framework for combining model predictions with observational data, accounting for uncertainties in both. It is particularly valuable for time-varying systems where states and parameters need to be estimated simultaneously.
The Ensemble Kalman Filter (EnKF) is a widely used DA method renowned for its capability in diverse domains such as atmospheric, oceanic, hydrologic, and biological systems [70]. EnKF works by running an ensemble of model realizations forward in time. When observations become available, the ensemble is updated (assimilated) based on the covariance between model states and the observations, thereby refining the estimates of both the current state and the model parameters [70].
Integrated Methodologies: ML-DA Synergy
The integration of ML and DA creates a powerful synergy for parameter estimation. ML can be used to pre-process data, learn non-linear relationships that inform observation operators, or emulate complex model components to speed up DA cycles. Conversely, DA can provide structured, uncertainty-aware frameworks for training ML models.
Table 1: ML-DA Integration Avenues for Land Surface Model Parameter Estimation [71] [72]
| Challenge in DA | Potential ML Solution |
|---|---|
| Identifying sensitive parameters and prior distributions | Use unsupervised ML for clustering and pattern detection to inform priors. |
| Characterizing model and observation errors | Train ML models on residuals to learn complex error structures. |
| Developing observation operators | Use neural networks to create non-linear operators linking model states to observations. |
| Handling large, heterogeneous datasets | Employ ML for efficient data reduction, feature extraction, and scaling. |
| Tackling spatial and temporal heterogeneity | Use spatially-aware ML (e.g., CNNs) or sequence models (e.g., RNNs) to handle context. |
One promising approach is hybrid modeling, which leverages the strengths of both model-based (physical) and data-driven (ML) modeling. For instance, in engineering, a hybrid model of a wind turbine blade bearing test bench used a physical finite element model combined with a Random Forest model to estimate non-measurable parameters like bolt preload. This hybrid approach improved the digital model's accuracy by up to 11%, enabling more effective virtual testing and condition monitoring [73].
Technical Implementation and Experimental Protocols
A Generic Workflow for ML-DA Parameter Estimation
The following diagram illustrates a consolidated workflow that integrates ML and DA for robust parameter estimation.
Detailed Methodological Breakdown
Phase 1: Problem Formulation and Data Preparation
The initial phase is critical for success and involves two key steps:
Step 1: Problem Scoping. Clearly define the model ( M ) and the set of parameters ( \theta ) to be estimated. Determine the state variables ( \textbf{x} ) and the available observations ( \textbf{y} ). Establish the dynamical rules (e.g., ODEs) and the known ranges or priors for the parameters.
Step 2: Data Curation. This is often the most time-consuming step, constituting up to 80% of the effort in an ML project [69]. The process involves:
- Collection: Gather high-dimensional data from all relevant sources (e.g., experimental readings, sensor data, omics data, images).
- Cleaning: Handle missing values, remove known outliers, and correct for systematic errors. Tools like Trifacta can be used for cleaning messy data [74].
- Harmonization: Map data from different sources to standard ontologies using tools like Tamr [74]. For legacy data trapped in silos, building a unified data platform on a system like a Cloudera Hadoop data lake may be necessary [74].
- Partitioning: Split the data into training, validation, and test sets. The validation set is used for tuning hyperparameters and the test set for a final, unbiased evaluation of the model's performance.
Phase 2: Model and Method Selection
Choosing the right combination of tools is paramount. The selection depends on the data type, problem context, and computational constraints.
Table 2: Machine Learning Toolbox for Parameter Estimation [69] [68]
| Method Category | Specific Algorithms/Architectures | Typical Application in Parameter Estimation |
|---|---|---|
| Supervised Learning | Random Forests, Gradient Boosting | Initial parameter estimation from features (e.g., bolt preload from strain gauges) [73]. |
| Deep Neural Networks (DNNs) | Fully Connected Feedforward Networks | Predictive model building with high-dimensional input (e.g., gene expression data) [69]. |
| Recurrent Neural Networks (RNNs, LSTM) | Analyzing time-series data where persistent information is needed [69]. | |
| Convolutional Neural Networks (CNNs) | Processing structured data (graphs) or image-based data (e.g., digital pathology) [69]. | |
| Deep Autoencoders (DAEN) | Unsupervised dimension reduction to preserve essential variables [69]. | |
| Data Assimilation | Ensemble Kalman Filter (EnKF) and variants | Refining parameter and state estimates by integrating observations with model forecasts [70]. |
Phase 3: Core Estimation and Validation
This phase executes the chosen methodology.
Protocol 1: ML-Based Estimation with Robust Loss. For neural network-based approaches, implement a robust training loop:
- Architecture: Design a network with appropriate layers (e.g., fully connected). The SiLU (Sigmoid Linear Unit) activation function has been shown to improve convergence in parameter estimation tasks [68].
- Loss Function: Employ the Huber loss to enhance robustness against noise and outliers. For a prediction error ( \delta ), the Huber loss ( L\delta ) is: [ L\delta(\delta) = \begin{cases} \frac{1}{2}{\delta^2} & \text{for } |\delta| \le \kappa, \ \kappa (|\delta| - \frac{1}{2}\kappa) & \text{otherwise.} \end{cases} ] where ( \kappa ) is a threshold parameter [68].
- Training: Use backpropagation and optimizers (e.g., Adam) to minimize the loss function, iteratively updating the network weights to learn the parameter values.
Protocol 2: Ensemble-Based DA Refinement. To incorporate the ML estimate into a dynamical system using DA:
- Initialization: Initialize an ensemble of model states, where the parameters of interest are part of the state vector. The initial parameter distribution can be informed by the ML estimate.
- Forecast Step: Propagate each ensemble member forward in time using the model ( M ).
- Analysis Step: When observations ( \textbf{y} ) are available, update each ensemble member using the EnKF equations: [ \thetai^a = \thetai^f + K (\textbf{y} - H(\textbf{x}i^f)) ] where ( \thetai^f ) and ( \theta_i^a ) are the parameter values for the ( i )-th member before and after assimilation, ( H ) is the observation operator, and ( K ) is the Kalman gain matrix, which weights the model forecast against the observations based on their relative uncertainties [70].
Protocol 3: Validation and UQ. Validate the final parameter set on the held-out test data. Use the ensemble spread from the DA cycle or statistical techniques like bootstrapping to quantify uncertainty in the parameter estimates. Metrics like root-mean-square error (RMSE) against a gold standard dataset are crucial for evaluating performance [69] [68].
The Scientist's Toolkit
Successful implementation of these advanced techniques requires a suite of computational tools and reagents.
Table 3: Essential Research Reagent Solutions for ML-DA Parameter Estimation
| Category / Item | Specific Examples | Function / Application |
|---|---|---|
| Programmatic Frameworks | TensorFlow, PyTorch, Scikit-learn [69] | Provides core libraries for building, training, and deploying machine learning models. |
| Data Processing Tools | StreamSets, Trifacta, Tamr [74] | Ingest, clean, and harmonize large, messy datasets from diverse sources. |
| Computing Infrastructure | Amazon SageMaker, Amazon EC2, GPUs/TPUs [69] [75] | Provides scalable, on-demand computing power for data-intensive modeling and training. |
| Data Assimilation Libraries | (e.g., DAPPER, PDAF) | Specialized software for implementing Ensemble Kalman Filters and other DA algorithms. |
| Data Storage | Amazon S3, Hadoop Data Lake [74] [75] | Scalable and secure storage for large volumes of structured and unstructured data. |
The integration of machine learning and data assimilation represents a paradigm shift in how researchers approach the fundamental problem of parameter estimation. ML provides the flexibility and power to learn from complex, high-dimensional data, while DA offers a rigorous, probabilistic framework for dynamically reconciling models with observations. The synergistic ML-DA methodology outlined in this guide provides a robust pathway to constrain model parameters more effectively, leading to enhanced model fidelity, reduced predictive uncertainty, and more reliable scientific insights. As data volumes continue to grow and models become more complex, this integrated approach will be indispensable for advancing research across the physical, biological, and engineering sciences.
Parameter estimation is a cornerstone of scientific computing, machine learning, and computational biology, enabling researchers to infer unknown model parameters from observational data [76]. However, real-world data is frequently contaminated by noise and incompleteness, which can severely distort traditional estimation methods. Noisy data contains errors, inconsistencies, or outliers that deviate from expected patterns, while incomplete data lacks certain values or observations [77] [78].
Traditional estimation approaches, particularly those based on least-squares criteria, exhibit high sensitivity to these data imperfections. Their quadratic loss functions disproportionately amplify the influence of outliers, leading to biased parameter estimates, reduced predictive accuracy, and ultimately, unreliable scientific conclusions and business decisions [79] [77]. Within life sciences and drug development, these inaccuracies can directly impact diagnostic models, therapeutic efficacy predictions, and clinical decision support systems.
This technical guide explores the formulation of robust cost functions and regularization techniques as a principled solution to these challenges. By designing objective functions that are less sensitive to data anomalies and that incorporate prior knowledge, researchers can develop estimation procedures that yield reliable, accurate, and interpretable models even with imperfect datasets. The content is framed within the broader thesis that careful mathematical formulation of the estimation problem itself is paramount for achieving robustness in the face of real-world data imperfections.
Foundations of Noisy and Incomplete Data
Characterizing Data Imperfections
Understanding the nature of data imperfections is the first step in selecting appropriate robustification strategies. Noise in quantitative research can be systematically categorized as shown in the table below.
Table 1: Taxonomy and Impact of Data Noise
| Noise Type | Description | Common Causes | Impact on Parameter Estimation |
|---|---|---|---|
| Random Noise | Small, unpredictable fluctuations around the true value. | Sensor imprecision, sampling errors, minor environmental variations [77]. | Increases variance of estimates but typically does not introduce bias under standard assumptions. |
| Systematic Noise | Consistent, predictable deviations from the true value. | Faulty instrument calibration, biased measurement protocols, persistent environmental factors [77]. | Introduces bias into parameter estimates, leading to consistently inaccurate models. |
| Outliers (Impulsive Noise) | Data points that lie an abnormal distance from other observations. | Sensor malfunction, data transmission errors, sudden cyberattacks, human data entry errors [79] [77]. | Can severely bias traditional least-squares estimators and distort the learned model structure. |
| Pure vs. Positive-Incentive Noise | Pure noise is detrimental; positive-incentive noise may contain useful latent information [80]. | Beneficial noise can arise from rare events or uncertainties that encourage model generalization [80]. | Pure noise degrades performance. Positive-incentive noise, if leveraged correctly, can potentially enhance robustness. |
The Problem with Traditional Least-Squares
The fundamental weakness of the conventional sum of squared-error criterion (SSEC) in the presence of outliers is its lack of bounded influence. Given a model error ( e(k) ), the SSEC utilizes a quadratic loss ( |e(k)|^2 ). When an outlier causes ( e(k) ) to be large, its squared value dominates the entire cost function ( J(\vartheta) = \sum_k |e(k)|^2 ). The optimization algorithm is then forced to adjust parameters ( \vartheta ) primarily to account for these few outliers, at the expense of fit quality for the majority of the data [79]. This can lead to significant performance deterioration and non-optimal models [79]. As noted in research on Errors-in-Variables (EIV) systems, this is particularly problematic when input as well as output data are contaminated by non-Gaussian noise [79].
A Taxonomy of Robust Cost Functions
Robust cost functions address the limitations of least-squares by reducing the sensitivity of the loss to large errors. The following table summarizes several key paradigms.
Table 2: Comparison of Robust Cost Function Paradigms
| Cost Function Paradigm | Mathematical Formulation | Robustness Mechanism | Best-Suited For | ||
|---|---|---|---|---|---|
| Continuous Mixed p-Norm (CMpN) | ( J1(\vartheta) := \int{1}^2 \lambda_k(p) \textrm{E}{ | e(k) | ^p} \textrm{d}p ) [79] | Averages various ( L_p )-norms (( 1 \leq p \leq 2 )), interpolating between L2 (sensitive) and L1 (robust) [79]. | Systems with impulsive noise where a single fixed norm is insufficient [79]. |
| Continuous Logarithmic Mixed p-Norm (CLMpN) | An enhanced version of CMpN designed to be differentiable and avoid stability issues [79]. | Uses a logarithmic transformation to improve differentiability and stability under impulsive noise [79]. | EIV nonlinear systems with aggressive impulsive noise requiring stable, differentiable optimization [79]. | ||
| M-Estimation & Information-Theoretic Learning | ( V(X,Y) = \textrm{E}[\kappa(X,Y)] ) (e.g., with Gaussian kernel ( G_\sigma(e) = \exp(-e^2/(2\sigma^2)) )) [81]. | Replaces quadratic loss with a kernel-based similarity measure (correntropy). Large errors are "down-weighted" due to the exponentially decaying kernel [81]. | Severe non-Gaussian noise, such as heavy-tailed distributions encountered in guidance systems [81]. | ||
| Dynamic Covariance Scaling (DCS) | ( \rho_S(\xi) = \frac{\theta \xi^2}{\theta + \xi^2} ) where ( \xi ) is the residual [81]. | A robust kernel where the cost saturates to ( \theta ) for large residuals, effectively nullifying the influence of extreme outliers [81]. | Real-time systems (e.g., robotics, guidance) where large, sporadic outliers are expected and computation is constrained [81]. |
Advanced Robust Formulations
Beyond the standard norms, recent research has developed more sophisticated frameworks. The Generalized M-Estimation-Based Framework combines M-estimation with information-theoretic learning to handle severe non-Gaussian noise in nonlinear systems, such as those found in guidance information extraction [81]. This hybrid approach uses a robust kernel function to down-weight the contribution of outliers during the state update process.
Another advanced concept challenges the notion that all noise is harmful. The Noise Tolerant Robust Feature Selection (NTRFS) method introduces the idea of "positive-incentive noise," suggesting that some noisy features can provide valuable information that encourages model generalization [80]. Instead of indiscriminately discarding noisy features, NTRFS employs ( \ell_{2,1} )-norm minimization and block-sparse projection learning to identify and exploit this beneficial noise, thereby enhancing robustness [80].
Regularization for Stability and Generalization
While robust cost functions handle outliers, regularization addresses the problem of model overfitting and ill-posedness, which can be exacerbated by noisy and incomplete data. Regularization incorporates additional information or constraints to stabilize the solution.
Sparsity-Inducing Regularization
A common assumption is that the underlying model or parameter vector is sparse, meaning only a few features or components are truly relevant. This is formalized using norm-based constraints.
- ( \ell1 )-Norm (Lasso): The ( \| \cdot \|1 ) promotes sparsity in the parameter vector by driving less important coefficients to exactly zero. It is the convex relaxation of the ideal but NP-hard ( \ell_0 )-norm problem [80].
- ( \ell_{2,1} )-Norm: This norm is particularly useful for group sparsity in multi-task or multi-class settings. It encourages entire rows of a parameter matrix to be zero, thereby selecting or discarding features jointly across all tasks [80].
- ( \ell_{2,0} )-Norm: This is the true sparsity measure, directly constraining the number of non-zero elements. While it is non-convex and NP-hard to optimize, recent methods like RPMFS and ESFS have developed practical algorithms, such as the Augmented Lagrangian Multiplier (ALM) method, to tackle it directly, eliminating the need for sensitive regularization parameter tuning [80].
Hybrid and Implicit Regularization
In complex scenarios like modeling partially known biological systems, Hybrid Neural Ordinary Differential Equations (HNODEs) offer a powerful form of implicit regularization [76]. In this framework, a neural network approximates unknown system dynamics, while a mechanistic ODE encodes known biological laws. The known physics acts as a structural regularizer, constraining the neural network from learning spurious patterns from the noisy data and guiding it towards physiologically plausible solutions [76].
Another implicit technique involves treating mechanistic parameters as hyperparameters during the training of an HNODE. This allows for a global exploration of the parameter space via hyperparameter tuning (e.g., using Bayesian Optimization), which helps avoid poor local minima that can trap standard gradient-based methods [76].
Experimental Protocols and Validation
Validating the performance of robust estimation methods requires rigorous experimental protocols. The following workflow, established in computational biology for HNODEs, provides a robust template for general parameter estimation problems [76].
Figure 1: Workflow for Robust Parameter Estimation and Identifiability Analysis
Step 1: Data Partitioning. The observed time-series data is split into training and validation sets. The training set is used for model calibration, while the validation set is held back to assess generalization performance and prevent overfitting [76].
Step 2a: Hyperparameter Tuning & Global Search. The incomplete model is embedded into a larger robust framework (e.g., HNODE, robust filter). Key parameters, including mechanistic parameters and regularization hyperparameters (e.g., kernel width ( \sigma ) in M-estimation, sparsity parameter ( \lambda )), are treated as hyperparameters. Global optimization techniques like Bayesian Optimization or genetic algorithms are employed to explore this combined search space, mitigating the risk of converging to poor local minima [76].
Step 2b: Model Training & Parameter Estimation. Using the promising initial estimates from Step 2a, the model is fully trained using a local, gradient-based optimizer (e.g., Adam, L-BFGS) to minimize the chosen robust cost function. This fine-tuning step refines the parameter estimates ( \hat{\boldsymbol{\theta}}^M ) [76].
Step 3: Practical Identifiability Analysis. After estimation, a practical identifiability analysis is conducted. This assesses whether the available data, with its inherent noise and limited observability, is sufficient to uniquely estimate the model parameters. This is typically done by analyzing the sensitivity of the cost function to perturbations in the parameter values or by examining the Fisher Information Matrix [76].
Step 4: Confidence Interval Estimation. For parameters deemed identifiable, asymptotic confidence intervals (CIs) are calculated to quantify the uncertainty in the estimates, providing a range of plausible values for each parameter [76].
The Scientist's Toolkit
Implementing the protocols above requires a set of essential computational and methodological "reagents." The following table outlines key components for a modern robust estimation pipeline.
Table 3: Research Reagent Solutions for Robust Estimation
| Reagent / Tool | Category | Function in Protocol |
|---|---|---|
| Bayesian Optimization | Global Optimization Algorithm | Efficiently explores the hyperparameter and mechanistic parameter space in Step 2a, balancing exploration and exploitation to find good initial estimates [76]. |
| Stochastic Approximation / SPSA | Optimization Algorithm | Tunes parameter vectors in Parameter-Modified Cost Function Approximations (CFAs) by evaluating performance over simulation trajectories, useful in Steps 2a and 2b [82]. |
| Augmented Lagrangian Multiplier (ALM) | Optimization Solver | Solves non-convex, constrained optimization problems, such as those involving ( \ell_{2,0} )-norm constraints for sparse feature selection [80]. |
| Hybrid Neural ODE (HNODE) | Modeling Framework | Embeds incomplete mechanistic knowledge into a differentiable model, serving as the core architecture for Steps 2a-2b when system dynamics are only partially known [76]. |
| Dynamic Covariance Scaling (DCS) | Robust Kernel Function | Used within a filter or optimizer to saturate the cost of large residuals, making the update step robust to outliers as in Step 2b [81]. |
| Implicit Function Theorem (IFT) | Analytical Tool | Enables dimensionality reduction in complex cost landscapes by profiling out parameters, simplifying the optimization in Step 2b [82]. |
Formulating a parameter estimation problem to be inherently robust is a critical step in ensuring the reliability of models derived from real-world, imperfect data. This guide has detailed how the choice of cost function—moving from traditional least-squares to robust paradigms like CLMpN, M-estimation, and DCS—directly controls the estimator's sensitivity to outliers. Furthermore, regularization techniques, particularly sparsity induction and physics-informed gray-box modeling, provide the necessary constraints to stabilize solutions and enhance generalization from incomplete data.
The presented experimental protocol and toolkit offer a structured approach for researchers, especially in drug development and computational biology, to implement these techniques. By systematically integrating robustness into the problem's foundation, scientists can produce parameter estimates that are not only statistically sound but also scientifically meaningful, thereby enabling more accurate predictions and trustworthy data-driven decisions.
Ensuring Reliability: Robust Validation and Comparative Analysis of Methods
In the rigorous field of data-driven research, particularly within scientific domains like drug development, the ability to accurately estimate model parameters and trust their performance on truly unseen data is paramount. The core challenge is overfitting—creating a model that excels on its training data but fails to generalize [83]. This paper frames cross-validation and back-testing not merely as evaluation tools, but as foundational components of a robust parameter estimation problem formulation. These methodologies provide the framework for objectively assessing a model's predictive power and ensuring that estimated parameters are meaningful and generalizable, rather than artifacts of a specific dataset [4] [9].
Core Concepts: From Overfitting to Generalization
The Fundamental Problem: Overfitting
In supervised machine learning, using the same data to both train a model and evaluate its performance constitutes a methodological error. A model could simply memorize the labels of the samples it has seen, achieving a perfect score yet failing to predict anything useful on new data. This situation is known as overfitting [83]. The standard practice to mitigate this is to hold out a portion of the available data as a test set (X_test, y_test).
The Role of a Validation Set
When manually tuning hyperparameters (e.g., the C setting in a Support Vector Machine), there remains a risk of overfitting to the test set because parameters can be tweaked until the estimator performs optimally. This leads to information leakage, where knowledge about the test set inadvertently influences the model, and evaluation metrics no longer reliably report generalization performance [83]. To address this, a validation set can be held out. Training proceeds on the training set, followed by evaluation on the validation set. Only after a successful experiment is the model evaluated on the final test set.
Cross-Validation as a Solution
Using a separate validation set reduces the data available for learning, and results can vary based on the random split. Cross-validation (CV) solves this problem. In the basic k-fold CV approach, the training set is split into k smaller sets (folds). The following procedure is repeated for each of the k folds:
- A model is trained using
k-1of the folds as training data. - The resulting model is validated on the remaining fold of data [83]. The reported performance measure is the average of the values computed across all loops. This approach is computationally expensive but does not waste data, which is a major advantage when samples are limited [83].
Methodological Deep Dive
Cross-Validation Techniques in Practice
The cross_val_score helper function in libraries like scikit-learn provides a simple way to perform cross-validation [83]. Beyond simple k-fold, other strategies include:
- ShuffleSplit: This method generates a predefined number of independent train/test dataset splits. Samples are first shuffled and then split into train/test sets [83].
- Stratified K-Fold: This ensures that each fold preserves the percentage of samples for each class, which is crucial for imbalanced datasets.
For more comprehensive evaluation, the cross_validate function allows for specifying multiple metrics and returns a dictionary containing fit-times, score-times, and optionally training scores and the fitted estimators [83].
The Critical Importance of Data Pipelines
A crucial best practice is to ensure that all data preprocessing steps (e.g., standardization, feature selection) are learned from the training set and applied to the held-out data. If these steps are applied to the entire dataset before splitting, information about the global distribution of the test set leaks into the training process [83]. Using a Pipeline is the recommended way to compose estimators and ensure this behavior under cross-validation, thereby preventing data leakage and providing a more reliable performance estimate [83].
Advanced Protocols: Back-Testing in Temporal Contexts
For data with temporal dependencies, such as financial returns or longitudinal clinical trials, standard cross-validation is problematic because it breaks the temporal order, potentially allowing future information to influence past predictions (look-ahead bias) [84].
Walk-Forward Back-Testing
This method respects temporal sequence:
- Train the model on an initial data segment up to time
t. - Test the model on the next subsequent block of time (e.g.,
t+1tot+n). - Expand the training set to include the previous test set.
- Repeat the process, "walking forward" in time until all data is used for testing [84].
Advantages: Eliminates look-ahead bias and naturally adapts to evolving data distributions. Drawbacks: Uses less training data initially and has a high overfitting risk if the model is over-optimized on small historical segments [84].
Comparative Analysis of Validation Protocols
The table below summarizes the key characteristics of different validation methodologies, aiding researchers in selecting the most appropriate protocol for their specific parameter estimation problem.
Table 1: Comparison of Model Validation Methodologies
| Protocol | Core Principle | Advantages | Limitations | Ideal Use Case |
|---|---|---|---|---|
| Holdout Validation | Single split into training and test sets. | Computationally simple and fast. | Results highly dependent on a single random split; inefficient data use. | Initial model prototyping with very large datasets. |
| K-Fold Cross-Validation [83] | Data partitioned into k folds; each fold serves as a test set once. |
Reduces variance of performance estimate; makes efficient use of data. | Susceptible to data leakage if not pipelined; problematic for temporal data. | Standard supervised learning on independent and identically distributed (IID) data. |
| Walk-Forward Back-Testing [84] | Sequential expansion of the training window with testing on the subsequent period. | Respects temporal order; no look-ahead bias; adapts to new data patterns. | Higher computational cost; less initial training data; risk of overfitting to local periods. | Financial modeling, clinical trial forecasting, and any time-series prediction. |
Experimental Framework & The Researcher's Toolkit
A Simulation-Based Case Study
To illustrate the dangers of overfitting, consider a simulation common in financial research but highly relevant to any field with high-dimensional data [84]:
- Objective: To demonstrate that seemingly strong model performance can arise by chance when testing many non-predictive features.
- Setup: Simulate autocorrelated asset returns. Introduce numerous random features (predictors) drawn from a normal distribution. By construction, these features are not predictive of returns.
- Protocol: Perform a 5-Fold Cross-Validation. In each run, select the best-performing factor based on in-sample correlation (IC) and compute the IC on the test fold.
- Results: After 1000 simulations, the distribution of Sharpe Ratios (a performance metric) showed a mean near zero. However, the scenario with "not many periods, more predictors" (T=1250, p=500) exhibited a significantly higher standard deviation, with some Sharpe Ratios exceeding 2.0 purely by chance [84]. This powerfully demonstrates that a high performance metric can be a spurious result of overfitting, not genuine predictive power.
The Scientist's Toolkit: Essential Research Reagents
For researchers implementing these protocols, the following tools are essential.
Table 2: Key Computational Tools for Model Validation
| Tool / Reagent | Function / Purpose | Technical Specification / Notes |
|---|---|---|
train_test_split [83] |
Helper function for quick random splitting of data into training and test subsets. | Critical for initial holdout validation. Requires careful setting of random_state for reproducibility. |
cross_val_score [83] |
Simplifies the process of running k-fold cross-validation for a single evaluation metric. | Returns an array of scores for each fold, allowing calculation of mean and standard deviation. |
cross_validate [83] |
Advanced function for multi-metric evaluation and retrieving fit/score times. | Essential for comprehensive model assessment and benchmarking computational efficiency. |
Pipeline [83] |
Chains together data preprocessors (e.g., StandardScaler) and final estimators. |
The primary tool for preventing data leakage during cross-validation, ensuring preprocessing is fit only on the training fold. |
| Walk-Forward Algorithm [84] | A custom implementation for time-series validation, respecting temporal order. | Requires careful design of the training window expansion logic and can be computationally intensive. |
Formulating a robust parameter estimation problem in research demands more than just sophisticated algorithms; it requires a rigorous framework for validation. Cross-validation provides a powerful standard for assessing generalization on IID data, while back-testing protocols like Walk-Forward are indispensable for temporal contexts. The simulation evidence clearly shows that without these safeguards, researchers are at high risk of mistaking overfitted, spurious patterns for genuine discovery. By integrating these methodologies into the core of the experimental design—using pipelines to prevent leakage and choosing validation strategies that match the data's structure—scientists and drug development professionals can produce models whose parameters are reliably estimated and whose performance on untested data can be trusted.
Parameter estimation is a cornerstone of empirical research across engineering, physical sciences, and life sciences. Formulating this problem effectively requires selecting an appropriate mathematical model and a robust algorithm to identify the model's unknown parameters from observed data [85] [86]. The core challenge lies in the optimization problem: minimizing the discrepancy between model predictions and experimental measurements. This guide, framed within a broader thesis on research methodology, examines a critical juncture in this formulation: the choice of optimization algorithm and the domain of analysis. Specifically, we benchmark three prominent algorithms—Particle Swarm Optimization (PSO), Grey Wolf Optimizer (GWO), and Least Squares (LSQ)—evaluating their performance in both the time and frequency domains [85]. The domain of analysis (time vs. frequency) fundamentally changes the data's representation and, consequently, the landscape of the optimization problem [87] [88]. A systematic comparison provides researchers with a evidence-based framework for aligning their algorithmic choice with their experimental data type and research objectives.
Core Algorithmic Methods and Experimental Domains
- Particle Swarm Optimization (PSO): A heuristic, population-based metaheuristic inspired by the social behavior of bird flocking. Particles (candidate solutions) fly through the search space, adjusting their velocity based on their own experience and the swarm's best-found position [89] [90]. It is known for rapid convergence but can be prone to premature convergence in local optima [90].
- Grey Wolf Optimizer (GWO): Another heuristic metaheuristic algorithm that simulates the social hierarchy and hunting mechanism of grey wolves. The population is categorized into alpha, beta, delta, and omega wolves, guiding the search process [89] [91]. It demonstrates strong exploration capabilities but may have weaker exploitation in later search stages [90].
- Least Squares (LSQ): A deterministic, local optimization method that finds the best-fitting parameters by minimizing the sum of the squares of the residuals (the differences between observed and predicted values). Its performance is highly dependent on a good initial guess and it is typically efficient for convex problems [89] [86].
Time Domain vs. Frequency Domain Analysis
The choice of analysis domain is not merely observational but transforms the nature of the parameter estimation problem.
- Time Domain Analysis: The signal or system response is analyzed as a function of time. This is intuitive for tracking transient responses, such as a battery's voltage drop under a load pulse or the vibrational decay of a structure [87] [92]. It provides direct insight into rise times, settling times, and overshoot [92].
- Frequency Domain Analysis: The signal is decomposed into its constituent sinusoidal frequencies, typically via a Fourier Transform [87] [88]. This reveals the power or amplitude at each frequency, simplifying the analysis of systems governed by linear differential equations and providing innate understanding of characteristics like bandwidth, resonant frequencies, and phase shift [87]. It is particularly powerful for analyzing periodic behaviors and noise components [93].
The mathematical transformation between domains means an algorithm may perform differently on the same underlying system when the data is presented in time versus frequency representations [85].
Detailed Experimental Protocols for Benchmarking
The following methodology is synthesized from comparative studies on battery equivalent circuit model (ECM) identification and nonlinear system identification [85] [86].
1. Problem Definition & Data Generation:
- System: Select a well-defined system with known ground-truth parameters. For example, a Thevenin equivalent circuit model for a lithium-ion battery (with parameters: open-circuit voltage, ohmic resistance, polarization resistance/time constant) [85] or a single-degree-of-freedom mechanical oscillator with a nonlinear stiffness element [86].
- Aging/Variation: To test robustness, data should be collected from the system under various states (e.g., different States of Health for batteries) [85].
- Measurement: Perform two parallel experimental campaigns:
- Time-domain: Apply a dynamic load profile (e.g., a current pulse or random excitation) and measure the voltage response over time.
- Frequency-domain: Perform Electrochemical Impedance Spectroscopy (EIS) or frequency sweep tests to obtain impedance spectra (magnitude and phase across a frequency range) [85].
2. Cost Function Formulation:
- Define a cost function, typically the Mean Squared Error (MSE) or Sum of Squared Errors (SSE), quantifying the difference between the measured data and the model's output [89] [86].
- For time-domain:
Cost = Σ(measured_voltage(t) - model_voltage(t))². - For frequency-domain:
Cost = Σ(|measured_impedance(f)| - |model_impedance(f)|)² + w·Σ(phase_error(f))², wherewis a weighting factor.
3. Algorithm Configuration & Execution:
- PSO: Set parameters: swarm size (e.g., 30-50), inertia weight (e.g., 0.7), cognitive and social coefficients (e.g., 1.5 each). Use adaptive strategies if needed [90].
- GWO: Set parameters: population size (e.g., 30-50), convergence factor
adecreasing linearly from 2 to 0. - LSQ: Use the Levenberg-Marquardt algorithm as a robust nonlinear least-squares solver. Provide a sensible initial parameter guess.
- Hybrid Methods (Optional): Implement hybrid starters, such as using PSO or GWO for global search to find a good initial point for LSQ [86].
- Run each algorithm on both the time-domain and frequency-domain datasets. Record the final cost value, computation time, and the identified parameters.
4. Performance Evaluation:
- Accuracy: Compare final parameter estimates against known ground truth.
- Consistency/Robustness: Run multiple trials (with different random seeds for PSO/GWO) to assess variance in results.
- Computational Efficiency: Compare the time or number of function evaluations to reach convergence.
- Convergence Behavior: Plot the cost function value versus iteration number to visualize convergence speed and stability.
Results and Comparative Analysis
The table below summarizes key findings from a comparative study on lithium-ion battery ECM parameter identification [85].
Table 1: Algorithm Performance in Time vs. Frequency Domains for ECM Parameter Identification
| Algorithm | Domain | Performance Summary | Key Strength | Key Limitation |
|---|---|---|---|---|
| PSO | Frequency | Optimal performance. Excels in navigating the frequency-domain error landscape. | Strong global search; avoids local minima. | Slower convergence than LSQ in time domain. |
| Time | Good, but often outperformed by LSQ. | Robust to initial guess. | Can be computationally intensive. | |
| GWO | Frequency | Optimal performance. Comparable to PSO, demonstrating effective exploration in frequency domain. | Good exploration-exploitation balance. | May require parameter tuning. |
| Time | Competitive, but not superior to LSQ. | Hierarchy-based search is effective. | Exploitation capability can be weaker [90]. | |
| LSQ | Frequency | Sub-optimal. Performance can degrade due to non-convexity of the frequency-domain cost function. | Very fast convergence when near optimum. | Highly dependent on an accurate initial guess. |
| Time | Superior performance. The time-domain cost function often favors the efficient local search of LSQ. | Computational efficiency and accuracy. | Prone to converging to local minima. |
General Conclusion: The study concluded that PSO and GWO are ideal candidates overall, with optimal performance in the frequency domain, while LSQ is superior in the time domain. This conclusion remained consistent across different battery aging states [85].
Advanced Insights: Hybridization and Robustness
- Hybrid Algorithms: To overcome individual limitations, hybrid methods are often employed. For instance, a global optimizer (PSO/GWO) can be used to find a promising region, after which a local method (LSQ) refines the solution, combining robustness with speed [86]. Other hybrids like DE-GWO (Differential Evolution-GWO) have been shown to improve global optimization capability and estimation accuracy over standard GWO [89].
- Model Complexity: Research indicates that when using superior identification methods like PSO or GWO, a simpler model structure can be sufficient and even beneficial for efficient State-of-Health monitoring, emphasizing the importance of the algorithm-model pairing [85].
Visualization of Research Workflow and Algorithm Performance
Diagram 1: Parameter Estimation Benchmarking Workflow
Diagram 2: Algorithm Performance in Time vs Frequency Domains
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 2: Key Research Reagents & Tools for Parameter Estimation Benchmarking
| Item Category | Specific Example/Name | Function in Research |
|---|---|---|
| Physical System under Test | Lithium-Ion Battery Pouch Cells | The device or material whose parameters (e.g., internal resistance, capacitance) are to be estimated. Aged under various conditions to test robustness [85]. |
| Data Acquisition Hardware | Bi-potentiostat / Frequency Response Analyzer | For electrochemical systems, applies controlled current/voltage excitations and measures high-fidelity voltage/current responses in both time and frequency domains [85]. |
| Signal Processing Tool | Fast Fourier Transform (FFT) Algorithm | Converts time-domain experimental data into the frequency-domain representation, enabling dual-domain analysis [87] [93]. |
| Optimization Software Library | SciPy (Python), Optimization Toolbox (MATLAB) | Provides implemented, tested versions of benchmark algorithms (PSO, Levenberg-Marquardt LSQ) and frameworks for coding custom algorithms like GWO. |
| Performance Metric | Mean Squared Error (MSE) / Sum of Squared Errors (SSE) | The quantitative cost function that algorithms minimize. Serves as the primary measure for comparing algorithm accuracy and convergence [89] [86]. |
| Validation Dataset | Prairie Grass Emission Experimental Data [89] or Synthetic Data with Known Truth | An independent, high-quality dataset used to validate the accuracy and generalizability of the parameters identified by the benchmarked algorithms. |
Formulating a parameter estimation problem requires a clear understanding of the relationship between the true goal—determining accurate parameter values—and the practical means of achieving it, which often involves minimizing model output error. This distinction is fundamental across scientific disciplines, particularly in drug development and systems biology, where mathematical models are used to describe complex biological systems. The core challenge lies in the inverse problem: predicting model parameters from observed data, a task for which a unique solution is often elusive [3]. A model's ability to describe data (output error) does not guarantee that the recovered parameters (parameter error) are correct or physiologically meaningful. This whitepaper provides a structured framework for analyzing these error metrics, enabling researchers to critically assess the reliability of their parameter estimates and thus the biological insights derived from them.
Theoretical Foundations: Error Types and Their Relationship
Defining Parameter Error and Model Output Error
In parameter estimation, two primary types of error are analyzed:
- Parameter Error: This quantifies the discrepancy between the estimated parameter values (( \hat{\theta} )) and their true, albeit often unknown, values (( \theta{true} )). It is defined as ( ||\hat{\theta} - \theta{true}|| ), where ( || \cdot || ) represents a suitable norm. Minimizing this error is the ultimate objective, as it ensures the model parameters reflect the underlying biology [3].
- Model Output Error (or Prediction Error): This is the discrepancy between the model's predictions (( y(\hat{\theta}) )) and the actual observed data (( Y )). It is frequently measured as the sum of squared residuals: ( \sum (Y - y(\hat{\theta}))^2 ). This error is the objective function minimized during the fitting process [3] [94].
The Critical Link: Practical Identifiability
A key challenge in parameter estimation is practical identifiability. A model may have low output error for a wide range of parameter combinations, leading to low parameter error in a structural identifiability sense, but high parameter error in practice if the available data is sparse or noisy [3]. This occurs because different parameter sets can produce nearly identical output trajectories, a phenomenon known as parameter correlation. Consequently, a well-fitted model (low output error) does not guarantee accurate parameters (low parameter error). The following workflow outlines the core process for evaluating these relationships.
Diagram 1: Core workflow for error analysis.
Methodologies for Error Analysis
Protocols for Evaluating Model Output Error
Evaluating model output error requires selecting appropriate metrics based on the model's purpose. The guiding principle is to use a strictly consistent scoring function for the target functional of the predictive distribution, as this ensures that minimizing the scoring function aligns with estimating the correct target [94].
Table 1: Common Metrics for Model Output Error
| Model Type | Target Functional | Strictly Consistent Scoring Function | Mathematical Form | Primary Use Case |
|---|---|---|---|---|
| Classification | Mean | Brier Score (Multiclass) | ( \frac{1}{N} \sum{i=1}^N \sum{j=1}^K (y{i,j} - p{i,j})^2 ) | Probability calibration assessment [94] |
| Classification | Mean | Log Loss | ( -\frac{1}{N} \sum{i=1}^N \sum{j=1}^K y{i,j} \log(p{i,j}) ) | Probabilistic prediction evaluation [94] |
| Regression | Mean | Squared Error | ( \frac{1}{N} \sum{i=1}^N (Yi - y(\hat{\theta})_i)^2 ) | Standard regression, assumes normal errors [94] |
| Regression | Quantile | Pinball Loss | ( \frac{1}{N} \sum{i=1}^N \begin{cases} \alpha (Yi - y(\hat{\theta})i), & \text{if } Yi \geq y(\hat{\theta})i \ (1-\alpha) (y(\hat{\theta})i - Y_i), & \text{otherwise} \end{cases} ) | Predicting specific percentiles/intervals [94] |
Experimental Protocol for Output Error Validation:
- Data Splitting: Partition the data into training and testing sets. The training set is used for parameter estimation, while the held-out testing set is used to compute the final output error, providing an estimate of model generalizability [95].
- Cross-Validation: For robust results, employ k-fold cross-validation, which involves repeating the train-test split multiple times to ensure the output error is stable and not dependent on a particular random data partition [95] [94].
- Metric Calculation: Compute the chosen strictly consistent scoring function (from Table 1) on the test set predictions. This quantifies the model's predictive performance on unseen data.
Protocols for Assessing Parameter Error and Identifiability
Since true parameters are typically unknown, parameter error is assessed indirectly through practical identifiability analysis. The following methods help determine if a unique set of parameters can be reliably estimated from the available data.
Table 2: Methods for Assessing Parameter Identifiability
| Method | Underlying Principle | Key Output | Interpretation |
|---|---|---|---|
| Correlation Matrix Analysis | Analyzes pairwise linear correlations between parameter sensitivities [3] | Correlation matrix | Parameters with correlations near ±1 indicate non-identifiability; one of them may be fixed. |
| Singular Value Decomposition (SVD) | Decomposes the sensitivity matrix to find orthogonal directions of parameter influence [3] | Singular values and vectors | Small singular values indicate poorly identifiable parameter combinations in the direction of the corresponding vector. |
| Subset Selection (QR) | Uses SVD followed by QR factorization with column pivoting to select a subset of identifiable parameters [3] | A subset of identifiable parameters | Provides a concrete set of parameters that can be estimated while others should be fixed. |
| Profile Likelihood | Systematically varies one parameter while re-optimizing others to explore the objective function shape [3] | Likelihood profiles for each parameter | Flat profiles indicate practical non-identifiability; well-formed minima suggest identifiability. |
Experimental Protocol for Parameter Identifiability:
- Compute Parameter Sensitivities: Calculate the matrix ( S ) of sensitivity coefficients, where each element ( S{ij} = \frac{\partial y(ti)}{\partial \theta_j} ), describing how the model output changes with small changes in each parameter [3].
- Apply Identification Method: Use one of the methods from Table 2, such as analyzing the correlation matrix derived from ( S ), to identify which parameters are highly correlated.
- Subset Selection and Re-estimation: Fix non-identifiable or highly correlated parameters to literature values and re-estimate the remaining subset. This reduces the problem's complexity and often leads to more reliable parameter estimates with lower error [3].
An Integrated Workflow for Error Comparison
The following workflow integrates the concepts of output error minimization and parameter identifiability assessment to guide the formulation of a robust parameter estimation problem. It highlights the iterative nature of model building and refinement.
Diagram 2: Integrated error evaluation workflow.
The Scientist's Toolkit: Key Reagents and Materials
Table 3: Essential Research Reagent Solutions for Parameter Estimation Studies
| Item / Reagent | Function in Analysis |
|---|---|
| Sensitivity Analysis Software (e.g., MATLAB, Python with SciPy) | Computes the partial derivatives of model outputs with respect to parameters, forming the basis for identifiability analysis [3]. |
| Global Optimization Toolbox (e.g., αBB, SCIP) | Solves the inverse problem by finding parameter sets that minimize output error, helping to avoid local minima which can distort error analysis [4]. |
| Strictly Consistent Scoring Functions (e.g., Brier Score, Pinball Loss) | Provides a truth serum for model evaluation, ensuring that the minimization of the score during parameter estimation leads to the correct statistical functional (e.g., mean, quantile) [94]. |
| Confusion Matrix & Derived Metrics (Precision, Recall, F1) | For classification models, decomposes output error into different types (false positives/negatives), allowing for cost-sensitive error analysis [95]. |
| Cross-Validation Framework (e.g., k-fold, LOO) | A statistical method used to assess how the results of a model will generalize to an independent dataset, providing a more reliable estimate of model output error [95] [94]. |
Formulating a robust parameter estimation problem requires a nuanced understanding of the interplay between parameter error and model output error. While minimizing output error is a necessary step, it is not sufficient for ensuring biologically accurate parameter estimates. Researchers must actively diagnose practical identifiability using structured methodologies such as correlation matrix analysis and subset selection. By integrating the assessment of parameter identifiability directly into the modeling workflow and using strictly consistent scoring functions for evaluation, scientists, especially in drug development, can make more reliable inferences about the physiological systems they study. This critical approach moves beyond mere curve-fitting to the principled estimation of meaningful biological parameters.
Parameter identification is a cornerstone of building quantitative, predictive mathematical models in computational biology. The process involves determining the unknown parameters of a model, such as kinetic rate constants and binding affinities, from experimental measurements of other quantities, like species concentrations over time [96]. These parameters are crucial for simulating system dynamics, testing biological hypotheses, and making reliable predictions about cellular behavior under new conditions. However, the parameter estimation problem is fraught with challenges. Biological models often contain tens to hundreds of unknown parameters, while experimental data are typically sparse and noisy, obtained from techniques like immunoblotting assays or fluorescent markers [97] [96]. Furthermore, the relationship between parameters and model outputs is often non-linear and can be insensitive to changes in parameter values, a property known as "sloppiness," making it difficult to identify unique parameter sets that fit the data [96].
The parameter estimation problem is formally defined as an optimization problem. Given a model that predicts outputs (\hat{y}(t)) and experimental data (y(t)), the goal is to find the parameter vector (\theta) that minimizes an objective function, most commonly a weighted residual sum of squares: (\sumi \omegai (yi - \hat{y}i)^2), where (\omegai) are weights, often chosen as (1/\sigmai^2) with (\sigmai^2) being the sample variance of the data point (yi) [97]. The reliability of the estimated parameters must then be evaluated through uncertainty quantification, which assesses how constrained the parameters are by the available data.
A Framework of Methodological Approaches
Parameter estimation methodologies can be broadly categorized into optimization techniques for finding point estimates and statistical methods for quantifying uncertainty.
Optimization Techniques for Point Estimation
Optimization algorithms are used to find the parameter values that minimize the chosen objective function. They can be divided into two main classes: gradient-based and gradient-free methods.
Table 1: Comparison of Optimization Methods for Parameter Estimation
| Method Class | Specific Algorithms | Key Principles | Advantages | Disadvantages |
|---|---|---|---|---|
| Gradient-Based | Levenberg-Marquardt, L-BFGS-B [97] | Utilizes gradient (and Hessian) of objective function to find local minima. | Fast convergence near optimum; efficient for high-dimensional problems [97]. | Can get stuck in local minima; requires gradient computation [97]. |
| Metaheuristic (Gradient-Free) | Young’s Double-Slit Experiment (YDSE), Gray Wolf Optimization (GWO), Differential Evolution [98] [99] | Uses a heuristic strategy to explore parameter space without derivatives. | Global search capability; resistant to local optima; problem-independent [98]. | Computationally expensive; no guarantee of optimality; may require many function evaluations [98]. |
| Hybrid | Cubic Regularized Newton with Affine Scaling (CRNAS) [100] | Combines second-order derivative information with regularization to handle constraints. | Focuses on points satisfying second-order optimality; handles constraints natively [100]. | Higher computational cost per iteration; complex implementation [100]. |
Gradient-based methods are efficient but require calculating the model's sensitivity to parameter changes. This can be done through several approaches:
- Finite Difference: A simple but inefficient and potentially inaccurate method for high-dimensional problems [97].
- Forward Sensitivity Analysis: Augments the original ordinary differential equation (ODE) system with equations for the derivatives of each species with respect to each parameter. This method is exact but becomes computationally expensive for models with many parameters and ODEs [97].
- Adjoint Sensitivity Analysis: A more complex method that drastically reduces computational cost for problems with many parameters by solving a single backward-in-time adjoint system [97]. It is particularly promising for large-scale biological models.
Metaheuristic algorithms have gained popularity for their ability to perform a global search. A recent novel algorithm, Young's Double-Slit Experiment (YDSE), was shown to outperform other metaheuristics like the Sine Cosine Algorithm and Gray Wolf Optimization in a parameter estimation problem for a proton exchange membrane fuel cell model, achieving a lower Sum of Square Error and faster convergence [98]. Hybrid methods, such as a Differential Evolution algorithm combined with the Nelder-Mead local search, have also been developed to balance global exploration and local refinement [99].
Uncertainty Quantification and Model Selection
After obtaining point estimates, it is crucial to quantify their uncertainty. This process determines the identifiability of parameters—whether the available data sufficiently constrain their possible values.
- Profile Likelihood: Involves varying one parameter and re-optimizing all others to find the confidence intervals for each parameter [97].
- Bootstrapping: Involves repeatedly fitting the model to resampled versions of the data to build an empirical distribution of the parameter estimates [97].
- Bayesian Inference: Estimates the full posterior probability distribution of the parameters, given the data and prior knowledge. This provides a complete picture of parametric uncertainty [97] [101].
Uncertainty quantification naturally leads to the problem of model selection, where multiple competing models of the same biological process are evaluated. Traditional methods like the Akaike Information Criterion (AIC) select a single "best" model. However, Bayesian Multimodel Inference (MMI) offers a powerful alternative that accounts for model uncertainty. Instead of choosing one model, MMI constructs a consensus prediction by taking a weighted average of the predictions from all candidate models: (\text{p}(q | d{\text{train}}, \mathfrak{M}K) = \sum{k=1}^{K} wk \text{p}(qk | \mathcal{M}k, d{\text{train}})), where (wk) are weights assigned to each model (\mathcal{M}_k) [101]. Methods for determining weights include Bayesian Model Averaging (BMA), pseudo-BMA, and stacking [101]. This approach increases the certainty and robustness of predictions, as demonstrated in a study of ERK signaling pathway models [101].
Experimental Protocols and Case Studies
Protocol: Optimal Experimental Design for PIP3 Signaling
A landmark study by Bandara et al. demonstrated the power of Optimal Experimental Design (OED) for parameter estimation in a live-cell signaling context [102]. The goal was to estimate parameters for a model of PI3K-induced production of the lipid phosphatidylinositol 3,4,5-trisphosphate (PIP3).
Experimental System:
- Inducible Recruitment System: A chemogenetic system using the rapamycin derivative iRap to recruit a cytosolic CF-p85 construct (CFP-FKBP12-p85 peptide) to the plasma membrane, activating endogenous PI3K.
- Readout: Translocation of a yellow fluorescent protein fused to the Akt PH domain (Y-PH) from the cytosol to the PIP3-enriched membrane.
- Inhibition: The PI3K inhibitor LY29 was used to perturb the system reversibly.
Optimal Design Workflow:
- Initial Intuitive Experiment: An intuitively designed experiment (e.g., a single step of iRap) was performed, but the derived data led to poorly constrained parameters.
- Parameter Uncertainty Prediction: A numerical algorithm was used to predict the uncertainty of parameter estimates for a prospective experimental protocol.
- Protocol Optimization: The algorithm calculated the concentration-time profiles for both iRap (inducer) and LY29 (inhibitor) that would minimize the predicted parameter uncertainty.
- Iterative Refinement: Two cycles of optimized experimentation and model refinement were sufficient to reduce the mean variance of parameter estimates by more than sixty-fold compared to the initial intuitive design [102].
This protocol highlights that optimally designed experiments, which strategically control input stimuli and sampling times, can dramatically improve parameter identifiability and minimize the number of required experiments.
Diagram 1: Optimal Experimental Design Workflow for PIP3 Signaling
Protocol: Data-Driven Model Selection for Pattern Formation
Hishinuma et al. developed a machine learning-based protocol for model selection and parameter estimation from static spatial pattern data, such as Turing patterns [103].
Workflow:
- Feature Extraction: A target pattern image is embedded into a 512-dimensional vector using a Vision Transformer (ViT) from a Contrastive Language-Image Pre-training (CLIP) model in a zero-shot setting.
- Model Selection: The similarity between the target image's vector and a database of vectors from patterns generated by various mathematical models (e.g., Turing, Gray-Scott, Eden) is computed. Models producing high-similarity patterns are selected as candidates.
- Parameter Estimation (Simulation-Decoupled Neural Posterior Estimation):
- The target image vector is dimensionally reduced.
- An approximate Bayesian inference is performed using Natural Gradient Boosting (NGBoost) to estimate the posterior distribution of the model parameters.
- Key Advantage: This method requires only a few steady-state images and does not need time-series data or initial conditions, making it highly applicable to biological imaging data [103].
Successfully implementing parameter estimation requires a suite of computational tools and an understanding of key experimental reagents.
Table 2: Key Software Tools for Parameter Estimation and Uncertainty Quantification
| Software Tool | Key Features | Applicable Model Formats |
|---|---|---|
| COPASI [97] | General-purpose software for simulation and analysis of biochemical networks. | SBML |
| Data2Dynamics [97] | Toolbox for parameter estimation in systems biology, focusing on dynamic models. | SBML |
| AMICI [97] | High-performance simulation and sensitivity analysis for ODE models. Used with PESTO for parameter estimation and UQ. | SBML |
| PyBioNetFit [97] | Parameter estimation tool with support for rule-based modeling and uncertainty analysis. | BNGL, SBML |
| Stan [97] | Statistical modeling platform supporting Bayesian inference with MCMC sampling and automatic differentiation. | ODEs, Statistical Models |
Research Reagent Solutions
- Fluorescent Protein Fusions (e.g., Y-PH): Used as live-cell biosensors to visualize and quantify the localization and dynamics of specific signaling molecules, such as PIP3 lipids [102].
- Chemogenetic Dimerization Systems (e.g., FKBP-FRB with iRap): Allow precise, rapid, and reversible control over protein localization and activity, enabling perturbation experiments critical for parameter estimation [102].
- Kinase Inhibitors (e.g., LY29): Small molecule inhibitors provide a means to perturb specific nodes in a signaling network, generating informative data for constraining model parameters [102].
Diagram 2: PIP3 Signaling Pathway for Parameter Estimation
This comparative analysis demonstrates that robust parameter identification requires a holistic strategy combining sophisticated computational methods with carefully designed experiments. Key takeaways include:
- No Single Best Algorithm: The choice between gradient-based, metaheuristic, and hybrid optimization methods depends on the problem's specific characteristics, including model size, non-linearity, and computational cost [97] [98] [100].
- Uncertainty is Non-Negotiable: Point estimates alone are insufficient; uncertainty quantification via profiling, bootstrapping, or Bayesian inference is essential for assessing the reliability of the model and its parameters [97] [101].
- Optimal Experiments are Transformative: Actively designing experiments using OED principles can yield dramatically more informative data than intuitive protocols, leading to better-constrained parameters with fewer experimental replicates [104] [102].
- Embrace Model Uncertainty: When multiple models are plausible, Bayesian Multimodel Inference provides a disciplined framework for making robust predictions that account for uncertainty in the model structure itself [101].
Future directions in the field point towards increased automation and integration. Machine learning, as seen in the automated model selection for spatial patterns, will play a larger role [103]. Furthermore, developing more efficient algorithms for handling large, multi-scale models and standardized workflows that seamlessly integrate experimental design, parameter estimation, and uncertainty quantification will be critical for advancing computational biology towards more predictive and reliable science.
In biomedical research, the ability to synthesize diverse evidence streams and accurately estimate model parameters is fundamental to advancing our understanding of complex biological systems, from molecular pathways to whole-organism physiology. Evidence synthesis provides structured methodologies for compiling and analyzing information from multiple sources to support healthcare decision-making [105]. This process systematically integrates findings from various study designs, enabling researchers to develop comprehensive insights that individual studies cannot provide alone. In computational biology, these synthesized insights often form the basis for mathematical models that describe biological processes, where parameter estimation emerges as a critical challenge [76]. Parameter estimation involves optimizing model parameters so that model dynamics align with experimental data, a process complicated by scarce or noisy data that often leads to non-identifiability issues where optimization problems lack unique solutions [76]. The integration of diverse evidence types—from randomized controlled trials to qualitative studies and after-action reports—with robust parameter estimation techniques represents a powerful approach for addressing complex biomedical questions, particularly in areas such as drug development, personalized medicine, and public health emergency preparedness [106] [107].
Frameworks for Evidence Synthesis in Biomedical Research
Evidence synthesis encompasses multiple methodological approaches, each designed to address specific types of research questions in biomedicine. Understanding these frameworks is essential for selecting appropriate methodology for specific biomedical applications.
Table 1: Common Evidence Synthesis Frameworks in Biomedical Research
| Framework | Discipline/Question Type | Key Components |
|---|---|---|
| PICO [108] | Clinical medicine | Patient, Intervention, Comparison, Outcome |
| PEO [108] | Qualitative research | Population, Exposure, Outcome |
| PICOT [108] | Education, health care | Patient, Intervention, Comparison, Outcome, Time |
| PICOS [108] | Medicine | Patient, Intervention, Comparison, Outcome, Study type |
| SPIDER [108] | Library and information sciences | Sample, Phenomenon of Interest, Design, Evaluation, Research type |
| CIMO [108] | Management, business, administration | Context, Intervention, Mechanisms, Outcomes |
The National Academies of Sciences, Engineering, and Medicine (NASEM) has developed an advanced framework specifically for synthesizing diverse evidence types in complex biomedical contexts. This approach adapts the GRADE (Grading of Recommendations, Assessment, Development, and Evaluations) methodology to integrate quantitative comparative studies, qualitative studies, mixed-methods studies, case reports, after-action reports, modeling studies, mechanistic evidence, and parallel evidence from analogous contexts [106]. This mixed-methods synthesis is particularly valuable in biomedical research where multiple dimensions of complexity exist, including intervention complexity, pathway complexity, population heterogeneity, and contextual factors [106]. The NASEM committee's 13-step consensus-building method involves comprehensive review of existing methods, expert consultation, and stakeholder engagement to develop a single certainty rating for evidence derived from diverse sources [106].
Parameter Estimation Methodologies in Computational Biology
Parameter estimation represents one of the central challenges in computational biology, where mathematical models are increasingly employed to study biological systems [76]. These models facilitate the creation of predictive tools and offer means to understand interactions among system variables [76].
Fundamental Approaches to Parameter Estimation
The well-established mechanistic modeling approach encodes known biological mechanisms into systems of ordinary or partial differential equations using kinetic laws such as mass action or Michaelis-Menten kinetics [76]. These equations incorporate unknown model parameters that must be estimated through optimization techniques that align model dynamics with experimental data. Established optimization methods include linear and nonlinear least squares, genetic and evolutionary algorithms, Bayesian optimization, control theory-derived approaches, and more recently, physics-informed neural networks [76].
Hybrid Modeling Frameworks
When mechanistic knowledge is incomplete, Hybrid Neural Ordinary Differential Equations (HNODEs) combine mechanistic ODE-based dynamics with neural network components [76]. Mathematically, HNODEs can be formulated as:
[\frac{d\mathbf{y}}{dt}(t) = f(\mathbf{y}, NN(\mathbf{y}), t, \boldsymbol{\theta}) \quad \mathbf{y}(0) = \mathbf{y_0}]
where (NN) denotes the neural network component, (f) encodes mechanistic knowledge, and (\theta) represents unknown mechanistic parameters [76]. This approach is also known as gray-box modeling or universal differential equations and has shown promise in computational biology applications [76].
Table 2: Parameter Estimation Methods in Computational Biology
| Method Category | Specific Techniques | Applications | Limitations |
|---|---|---|---|
| Traditional Optimization | Linear/nonlinear least squares, Genetic algorithms, Bayesian Optimization | Well-characterized biological systems with complete mechanistic knowledge | Requires detailed understanding of system interactions; struggles with partially known systems |
| Hybrid Approaches | HNODEs, Gray-box modeling, Universal differential equations | Systems with partially known mechanisms; multi-scale processes | Potential parameter identifiability issues; requires specialized training approaches |
| Artificial Intelligence | Physics-informed neural networks, AI-mechanistic model integration | Complex systems with multi-omics data; drug discovery; personalized medicine | Limited interpretability; high computational requirements |
Recent advances integrate artificial intelligence (AI) with mechanistic modeling to address limitations of both approaches [107]. While AI can integrate multi-omics data to create predictive models, it often lacks interpretability, whereas mechanistic modeling produces interpretable models but struggles with scalability and parameter estimation [107]. The integration of these approaches facilitates biological discoveries and advances understanding of disease mechanisms, drug development, and personalized medicine [107].
Experimental Protocols and Workflow Integration
End-to-End Parameter Estimation and Identifiability Analysis
For scenarios with incomplete mechanistic knowledge, a robust workflow for parameter estimation and identifiability analysis involves multiple stages [76]:
- Data Preparation: Split experimental observation time points into training and validation sets, focusing on observable variables within the system.
- Model Expansion: Embed the incomplete mechanistic model into a HNODE framework, using Bayesian Optimization to simultaneously tune model hyperparameters and explore the mechanistic parameter search space.
- Model Training: Fully train the HNODE model to yield mechanistic parameter estimates using gradient-based methods.
- Identifiability Analysis: Assess local identifiability at-a-point of the parameters, extending well-established methods for mechanistic models to the HNODE framework.
- Confidence Interval Estimation: For locally identifiable parameters, estimate asymptotic confidence intervals to quantify uncertainty [76].
This workflow has been validated across various in silico scenarios, including the Lotka-Volterra model for predator-prey interactions, cell apoptosis models with inherent non-identifiability, and oscillatory yeast glycolysis models [76].
Evidence Synthesis Methodology
The NASEM methodology for evidence synthesis in public health emergency preparedness and response provides a transferable framework for biomedical applications [106]. This mixed-methods approach involves:
- Conducting method-specific reviews (systematic reviews, qualitative evidence syntheses, etc.) for each evidence stream.
- Assessing certainty and confidence in evidence using adapted GRADE methodology.
- Synthesizing findings across evidence streams through mapping qualitative to quantitative findings.
- Developing a single certainty rating for mixed-methods evidence to inform decision-making [106].
This methodology is particularly valuable for complex biomedical questions involving multiple evidence types, such as clinical trials, real-world evidence, patient preferences, and economic considerations.
Figure 1: Workflow for parameter estimation and identifiability analysis with incomplete mechanistic knowledge, adapting methodologies from computational biology [76].
Visualization Standards for Biomedical Research Communication
Effective data visualization plays a crucial role in communicating complex biomedical research findings. The strategic use of color palettes enhances comprehension, supports accessibility, establishes hierarchy, and creates visual appeal [109]. Visualization design should follow a structured process: (1) identify the core message, (2) describe the visualization approach, (3) create a draft visualization, and (4) fine-tune the details [110].
Color Palette Specifications for Biomedical Visualizations
Based on evidence from data visualization research, the following color specifications ensure clarity and accessibility:
- Sequential Palettes: Use for numerical representations with clear hierarchy or progression (e.g., heat maps, temperature variations) [109]
- Qualitative Palettes: Apply for categorical variables without inherent order (e.g., cell types, treatment groups), limiting to 10 or fewer colors [109]
- Diverging Palettes: Implement to emphasize contrast between two data segments with a meaningful midpoint (e.g., upregulated/downregulated genes) [109]
Accessibility considerations are paramount, affecting over 4% of the population with color vision deficiencies. High-contrast combinations and readability-enhancing palettes promote inclusivity [109]. The specified color palette (#4285F4, #EA4335, #FBBC05, #34A853, #FFFFFF, #F1F3F4, #202124, #5F6368) provides sufficient contrast when combinations are carefully selected.
Figure 2: Framework for synthesizing diverse evidence streams to develop a single certainty rating, adapting the NASEM methodology for biomedical applications [106].
Table 3: Essential Research Reagent Solutions for Biomedical Parameter Estimation
| Reagent/Resource | Function | Application Context |
|---|---|---|
| Hybrid Neural ODE (HNODE) Framework [76] | Combines mechanistic modeling with neural networks to represent unknown system components | Parameter estimation with incomplete mechanistic knowledge |
| Bayesian Optimization Tools [76] | Global exploration of mechanistic parameter space during hyperparameter tuning | Optimization in complex parameter landscapes with multiple local minima |
| Identifiability Analysis Methods [76] | Assess structural and practical identifiability of parameters post-estimation | Evaluating reliability of parameter estimates from scarce or noisy data |
| GRADE-CERQual Methodology [106] | Assesses confidence in qualitative evidence syntheses | Qualitative and mixed-methods evidence evaluation |
| AI-Mechanistic Integration Platforms [107] | Integrates multi-omics data with interpretable mechanistic models | Biological discovery, drug development, personalized medicine |
| Digital Twin Technologies [107] | Creates virtual patient models for pharmacological discoveries | Drug development, treatment optimization, clinical trial design |
The integration of robust evidence synthesis methodologies with advanced parameter estimation techniques represents a powerful paradigm for addressing complex challenges in biomedical research. The frameworks and protocols outlined in this technical guide provide researchers with comprehensive approaches for generating reliable, actionable insights from diverse evidence streams. As biomedical questions grow increasingly complex—spanning multiple biological scales, evidence types, and methodological approaches—the ability to synthesize results and make informed methodological choices becomes ever more critical. By adopting these structured approaches, researchers and drug development professionals can enhance the rigor, reproducibility, and impact of their work, ultimately accelerating progress toward improved human health outcomes.
Conclusion
Formulating a parameter estimation problem is a systematic process that moves from abstract concepts to a concrete, solvable optimization framework. Success hinges on a clear definition of parameters and models, a careful formulation of the objective function and constraints, and the strategic application of advanced optimization and troubleshooting techniques for complex biomedical systems. Looking forward, the integration of machine learning with traditional data assimilation, along with the development of more efficient global optimization methods like hybrid and graph-based strategies, promises to significantly reduce uncertainties in critical areas such as drug development and clinical trial modeling. By adopting this comprehensive framework, researchers can enhance the predictive power of their models, leading to more reliable and impactful scientific outcomes.
References
- 1. What are parameters, parameter estimates, and sampling ... [https://support.minitab.com/en-us/minitab/help-and-how-to/statistics/basic-statistics/supporting-topics/data-concepts/what-are-parameters-parameter-estimates-and-sampling-distributions/]
- 2. Parameter - Overview, Examples, and Uses in Statistics [https://corporatefinanceinstitute.com/resources/data-science/parameter/]
- 3. A practical approach to parameter estimation applied ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3526689/]
- 4. Parameter estimation in nonlinear algebraic models via ... [https://www.sciencedirect.com/science/article/pii/S0098135498002178]
- 5. Population Parameter - Lean Six Sigma Glossary Term [https://sixsigmadsi.com/glossary/population-parameter/]
- 6. Parameter vs Statistic: Examples & Differences [https://statisticsbyjim.com/basics/parameter-vs-statistic/]
- 7. Population Parameter - an overview [https://www.sciencedirect.com/topics/social-sciences/population-parameter]
- 8. Parameter - Learn Statistics [https://statdictionary.com/basic_stat_terms/p/parameter/]
- 9. Formulation of a practical algorithm for parameter ... [https://ntrs.nasa.gov/citations/19840035824]
- 10. Advancing drug development with “Fit-for-Purpose” modeling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12436579/]
- 11. Bayesian and subset-selection methods for parameter ... [https://www.sciencedirect.com/science/article/pii/S0263876225001017]
- 12. Chapter 10 Contrasts | Analysing Data using Linear Models [https://bookdown.org/pingapang9/linear_models_bookdown/contrasts.html]
- 13. Editorial: Application of PKPD modeling in drug discovery ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1614689/full]
- 14. Mass spectrometry methods and mathematical PK/PD ... [https://www.nature.com/articles/s41467-025-56985-6]
- 15. A review on model-based design of experiments for ... [https://www.sciencedirect.com/science/article/pii/S0009250925011686]
- 16. 6 Data Quality Metrics and Key Measures for Reliable Data [https://firsteigen.com/blog/6-key-data-quality-metrics-you-should-be-tracking/]
- 17. The 6 Data Quality Dimensions with Examples [https://www.collibra.com/blog/the-6-dimensions-of-data-quality]
- 18. 12 Data Quality Metrics to Measure Data Quality in 2025 [https://lakefs.io/data-quality/data-quality-metrics/]
- 19. 4 Types of Data - Nominal, Ordinal, Discrete, Continuous [https://www.mygreatlearning.com/blog/types-of-data/]
- 20. Types of Variables in Research & Statistics | Examples [https://www.scribbr.com/methodology/types-of-variables/]
- 21. Focus on Data: Statistical Design of Experiments and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7425741/]
- 22. Group and sample size | NC3Rs EDA [https://eda.nc3rs.org.uk/experimental-design-group]
- 23. Difference between Point and Interval Estimate [https://www.geeksforgeeks.org/maths/difference-between-point-and-interval-estimate/]
- 24. Point estimation [https://en.wikipedia.org/wiki/Point_estimation]
- 25. What's the difference between a point estimate and an ... [https://www.scribbr.com/frequently-asked-questions/point-estimate-vs-interval-estimate/]
- 26. What's the advantage of a point estimate over an interval ... [https://stats.stackexchange.com/questions/493296/whats-the-advantage-of-a-point-estimate-over-an-interval-estimate]
- 27. Point and Interval Estimation [https://sixsigmastudyguide.com/point-and-interval-estimation/]
- 28. MODEL PARAMETERS AND ASSUMPTIONS - Drug ... - NCBI [https://www.ncbi.nlm.nih.gov/books/NBK611884/]
- 29. Lesson 1: Point Estimation | STAT 415 [https://online.stat.psu.edu/stat415/lesson/1]
- 30. The distinction between confidence intervals, prediction ... [https://www.graphpad.com/support/faq/the-distinction-between-confidence-intervals-prediction-intervals-and-tolerance-intervals/]
- 31. A deep dive into parameter estimation for advanced PBPK ... [https://www.sciencedirect.com/science/article/pii/S134743672400017X]
- 32. Automated drug design for druggable target identification ... [https://www.nature.com/articles/s41598-025-18091-x]
- 33. How the Software Formulates Parameter Estimation as an ... [https://www.mathworks.com/help/sldo/ug/optimization-problem-formulation-for-parameter-estimation.html]
- 34. Loss Functions & the Sum of Squared Errors Loss [https://dustinstansbury.github.io/theclevermachine/cutting-your-losses]
- 35. Why use MSE instead of SSE as cost function in linear ... [https://stats.stackexchange.com/questions/539137/why-use-mse-instead-of-sse-as-cost-function-in-linear-regression]
- 36. SAE - Sum of Absolute Errors – Help center [https://support.numxl.com/hc/en-us/articles/214286506-SAE-Sum-of-Absolute-Errors]
- 37. The difference of SSE and MAE curve fitting and optimization [https://www.mathworks.com/matlabcentral/answers/1714600-the-difference-of-sse-and-mae-curve-fitting-and-optimization]
- 38. 8.2.3 Maximum Likelihood Estimation [https://www.probabilitycourse.com/chapter8/8_2_3_max_likelihood_estimation.php]
- 39. Maximum likelihood estimation [https://en.wikipedia.org/wiki/Maximum_likelihood_estimation]
- 40. Inequality constrained parameter estimation using filtering ... [https://www.sciencedirect.com/science/article/abs/pii/S0009250913007744]
- 41. fitting PBPK models to observed clinical data [https://pmc.ncbi.nlm.nih.gov/articles/PMC4294076/]
- 42. Model-Based MR Parameter Mapping with Sparsity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4152400/]
- 43. lsqnonlin - Solve nonlinear least-squares ... [https://www.mathworks.com/help/optim/ug/lsqnonlin.html]
- 44. lsqnonlin (Optimization Toolbox) [http://www.ece.northwestern.edu/local-apps/matlabhelp/toolbox/optim/lsqnonlin.html]
- 45. fmincon - Find minimum of constrained nonlinear ... [https://www.mathworks.com/help/optim/ug/fmincon.html]
- 46. Particle swarm optimization [https://en.wikipedia.org/wiki/Particle_swarm_optimization]
- 47. Compare lsqnonlin and fmincon for Constrained Nonlinear ... [https://www.mathworks.com/help/optim/ug/compare-lsqnonlin-fmincon.html]
- 48. Code Generation in fmincon Background - MATLAB & ... [https://www.mathworks.com/help/optim/ug/code-generation-in-fmincon.html]
- 49. Implementation of Matlab's fmincon function in C++ [https://stackoverflow.com/questions/5439321/implementation-of-matlabs-fmincon-function-in-c]
- 50. Cosmological Parameter Estimation using Particle Swarm ... [https://arxiv.org/abs/2508.05757]
- 51. Frontiers | A novel method to estimate the absorption rate constant... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2023.1087913/full]
- 52. Overview of oral absorption models and modelling issues [https://www.page-meeting.org/Abstracts/overview-of-oral-absorption-models-and-modelling-issues/]
- 53. Drug Absorption Modeling as a Tool to Define the Strategy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2621306/]
- 54. Model of double absorption of veralipride [https://monolixsuite.slp-software.com/tutorials/2024R1/case-study-model-of-double-absorption-of-veralipri]
- 55. Implementation of a transit compartment model for describing drug ... [https://link.springer.com/article/10.1007/s10928-007-9066-0]
- 56. From preclinical to human--prediction of oral absorption and... [https://pubmed.ncbi.nlm.nih.gov/22383166/]
- 57. Summary of the DREAM8 Parameter Estimation Challenge [https://pmc.ncbi.nlm.nih.gov/articles/PMC4447414/]
- 58. Nonidentifiability in model calibration and implications for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6156799/]
- 59. Predictive power of non-identifiable models [https://www.nature.com/articles/s41598-023-37939-8]
- 60. How do we Deal with Identifiability Problems in Statistics? [https://stats.stackexchange.com/questions/620217/how-do-we-deal-with-identifiability-problems-in-statistics]
- 61. Parameter correlations while curve fitting [https://www.sciencedirect.com/science/article/pii/S007668790021207X]
- 62. An optimal experimental design strategy for improving ... [https://www.sciencedirect.com/science/article/pii/S0098135423000029]
- 63. Optimal Experimental Design Based on Two-Dimensional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8906444/]
- 64. Optimal experimental design for parameter estimation in ... [https://arxiv.org/abs/2504.19233]
- 65. Clinical Trial Simulations [https://sph.uth.edu/dept/bads/faculty-home/hulinwu/research-projects/simulations]
- 66. How to Write a Research Protocol: Tips and Tricks - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6172884/]
- 67. How to write a lab protocol [https://www.scinote.net/blog/how-to-structure-a-protocol/]
- 68. Machine learning in parameter estimation of nonlinear ... [https://link.springer.com/article/10.1140/epjb/s10051-025-00904-7]
- 69. Applications of machine learning in drug discovery and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6552674/]
- 70. 20th international EnKF workshop (2025) [https://www.data-assimilation.no/workshops/EnKF-WS-2025]
- 71. Raoult et al. 2025: Parameter estimation in land surface ... [https://www.giss.nasa.gov/pubs/abs/ra09110p.html]
- 72. Parameter estimation in land surface models: challenges and ... [https://nora.nerc.ac.uk/id/eprint/540472]
- 73. Machine learning based parameter estimation for an ... [https://www.sciencedirect.com/science/article/pii/S2666546824001022]
- 74. Big Data in Pharma: Case Studies from Drug Discovery to ... [https://intuitionlabs.ai/articles/big-data-case-studies]
- 75. Insilico Case Study | Life Sciences [https://aws.amazon.com/solutions/case-studies/insilico-case-study/]
- 76. Robust parameter estimation and identifiability analysis ... [https://www.nature.com/articles/s41540-024-00460-3]
- 77. Key Techniques for Dealing with Noisy Data [https://www.sapien.io/blog/key-techniques-for-dealing-with-noisy-data]
- 78. A Primer of Data Cleaning in Quantitative Research [https://pubmed.ncbi.nlm.nih.gov/40145308/]
- 79. Robust recursive estimation for the errors-in-variables ... [https://www.nature.com/articles/s41598-025-89969-z]
- 80. Improve noise tolerance of robust feature selection via ... [https://www.sciencedirect.com/science/article/abs/pii/S0031320325006880]
- 81. Generalized M-Estimation-Based Framework for Robust ... [https://www.mdpi.com/1099-4300/27/12/1217]
- 82. Parameter-Modified Cost Function [https://www.emergentmind.com/topics/parameter-modified-cost-function]
- 83. 3.1. Cross-validation: evaluating estimator performance [https://scikit-learn.org/stable/modules/cross_validation.html]
- 84. Backtesting Series – Episode 2: Cross-Validation techniques [https://bsic.it/backtesting-series-episode-2-cross-validation-techniques/]
- 85. A comparative study of parameter identification methods for ... [https://www.sciencedirect.com/science/article/pii/S2352152X25004207]
- 86. Benchmarking Optimisation Methods for Model Selection ... [https://www.mdpi.com/2571-631X/4/3/36]
- 87. Time Domain Analysis vs Frequency ... [https://resources.pcb.cadence.com/blog/2020-time-domain-analysis-vs-frequency-domain-analysis-a-guide-and-comparison]
- 88. What's The Difference Between Time Domain and ... [https://maurymw.com/whats-the-difference-between-time-domain-and-frequency-domain-analysis/?srsltid=AfmBOorpwDswnQSL6UGZeZmb2gWjyh0C3x0ldUky7aaVaWJeKSkpGFm4]
- 89. An improved grey wolf optimizer algorithm for identification ... [https://www.sciencedirect.com/science/article/abs/pii/S0950423023000335]
- 90. An intelligent hybrid grey wolf-particle swarm optimizer for ... [https://www.nature.com/articles/s41598-025-02154-0]
- 91. Prediction of solar irradiance using grey wolf Optimizer-Least ... [https://ijeecs.iaescore.com/index.php/IJEECS/article/view/20574]
- 92. Time domain vs Frequency domain Analysis: What, When ... [https://medium.com/@svm161265/time-domain-vs-frequency-domain-analysis-what-when-and-whys-1e8f91fc3043]
- 93. A Robust High Precision Algorithm for Sinewave Parameter ... [https://pdxscholar.library.pdx.edu/open_access_etds/4685/]
- 94. 3.4. Metrics and scoring: quantifying the quality of predictions [https://scikit-learn.org/stable/modules/model_evaluation.html]
- 95. 12 Important Model Evaluation Metrics for Machine ... [https://www.analyticsvidhya.com/blog/2019/08/11-important-model-evaluation-error-metrics/]
- 96. Parameter Estimation and Model Selection in Computational ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1000696]
- 97. Parameter Estimation and Uncertainty Quantification for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7384601/]
- 98. An efficient approach for mathematical modeling and ... [https://www.nature.com/articles/s41598-025-10394-3]
- 99. Comparing optimization algorithms for parameter ... [https://link.springer.com/article/10.1007/s40430-024-04698-0]
- 100. Novel Optimization Techniques for Parameter Estimation [https://arxiv.org/abs/2407.04235]
- 101. Increasing certainty in systems biology models using ... [https://www.nature.com/articles/s41467-025-62415-4]
- 102. Optimal Experimental Design for Parameter Estimation of a ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1000558]
- 103. Data-driven discovery and parameter estimation of ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1012689]
- 104. Optimal experimental design for parameter estimation in ... [https://arxiv.org/html/2504.19233v1]
- 105. Overview of evidence synthesis types and modes [https://www.sciencedirect.com/science/article/pii/S0895435625003038]
- 106. A framework for synthesizing intervention evidence from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10084284/]
- 107. Artificial-intelligence-driven Innovations in Mechanistic ... [https://pubmed.ncbi.nlm.nih.gov/40316010/]
- 108. Guides: Evidence Synthesis: Frameworks [https://libguides.baylor.edu/evidencesynthesis/frameworks]
- 109. Mastering The Art of Data Visualization Color Palettes [https://www.datylon.com/blog/a-guide-to-data-visualization-color-palette]
- 110. Data Visualization: Rules and Guidelines [https://datascience.quantecon.org/tools/visualization_rules.html]